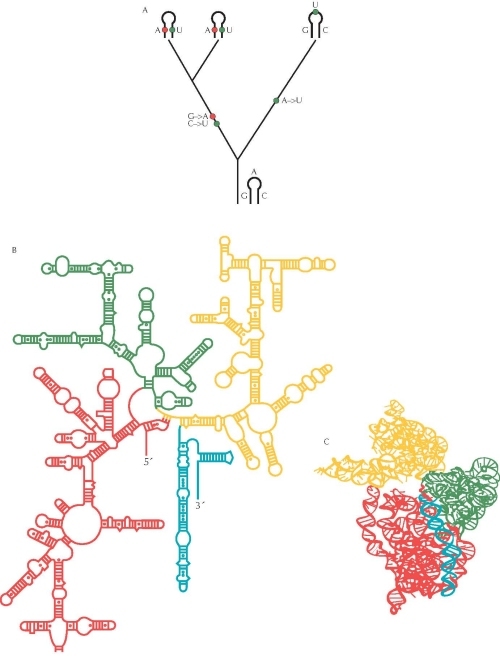

Figure 3. The pattern of evolutionary change in an RNA molecule can be used to determine its structure. (A) Where two parts of the RNA molecule pair, a change in one base must be accompanied by a change in its partner if pairing is to be maintained. In this example, G is replaced by A (red) and C by U (blue), in the common ancestor of two of the species. This maintains the pairing between these bases. In contrast, changes in unpaired regions can occur independently. In this example, there is a single change from A to U in the ancestry of the third species (green). (B) The secondary structure of the 16S ribosomal RNA molecule was inferred using this method, based on comparisons between more than 7000 sequences from different bacterial species. Ladders indicate paired helices. (C) The full three-dimensional structure of crystals of the molecule was determined directly in 2000 and confirms the inference made by the comparative method: More than 97% of the predicted pairs were found in the crystal structure. Different colored regions in B and C correspond to different domains of the molecule. Source: B and C, Adapted from Wimberly B.T. et al. 2000. Structure of the 30S ribosomal subunit. Nature 407: 327–339 (Fig. 2a,b, p. 329). http://www.nature.com