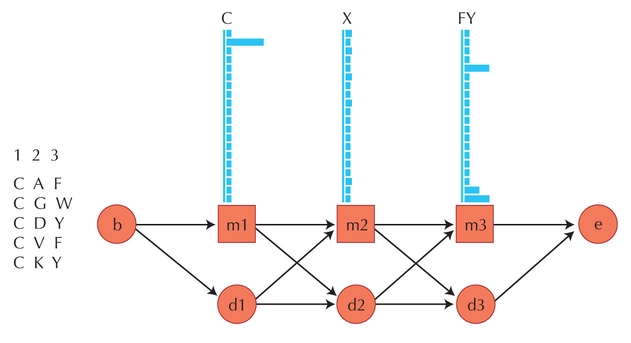

Figure 27.7. A simple hidden Markov model for a sequence alignment. (Left) An alignment is shown for five related peptides, each with three amino acids. These three columns are used to create a model of the sequence patterns of this peptide group. (Right) Hidden Markov model for these five peptides. The columns are modeled by the match states (boxes labeled m1, m2, and m3), each of which has 20 possible residues with the probability for each determined from the alignment column. (b and e circles) Beginning and ending probabilities. The arrows represent transition probabilities between states. (Modified from Eddy S.R. Bioinfo. Rev. 14: 755–763, Fig. 2, © 1998 Oxford University Press.)
| © 2007-2010 by Cold Spring Harbor Laboratory Press. All rights reserved. |
| The reproduction, modification, storage in a retrieval system, or retransmission, in any form or by any means, electronic, mechanical, or otherwise, for reasons other than personal, noncommercial use is strictly prohibited without prior written permission. You are authorized to download one copy of the material on this Web site for personal, noncommercial use only. The material made available on this Web site is protected by United States copyright laws and is provided solely for the use of instructors in teaching their courses and assessing student learning. Dissemination or sale of any of this material, as a whole or in parts (including on the World Wide Web), is not permitted. All users of these materials and visitors to this Web site are expected to abide by these restrictions. Requests for permission for other uses of this material should be directed to Cold Spring Harbor Laboratory Press, 1 Bungtown Road, Cold Spring Harbor, NY 11724 or submitted via our World Wide Web Site at http://www.cshlpress.com/. |