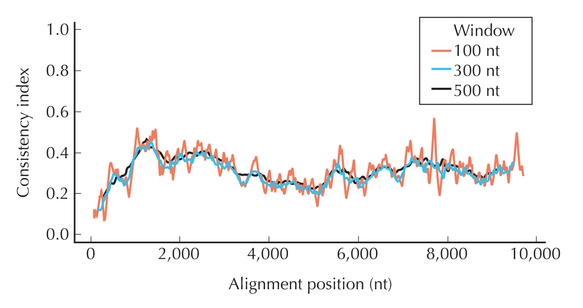

Figure 27.22. Consistency index (CI). A CI score has been plotted for the whole genome of hepatitis C viruses. First, an alignment of whole genomes of different hepatitis C viruses was made. Then a phylogenetic tree was inferred for the complete alignment. Third, small regions of the whole alignment were analyzed to determine how consistent the data in that region was relative to the tree of the whole genome. A sliding window of different base pair lengths was used and the CI for each position in an alignment of complete genomes was averaged over this window. Window sizes are 100 (red), 300 (blue), or 500 (black) nucleotides. (Redrawn from Hraber P.T. et al. 2006. Virol. J. 3: 103, Fig. 3a.)
| © 2007-2010 by Cold Spring Harbor Laboratory Press. All rights reserved. |
| The reproduction, modification, storage in a retrieval system, or retransmission, in any form or by any means, electronic, mechanical, or otherwise, for reasons other than personal, noncommercial use is strictly prohibited without prior written permission. You are authorized to download one copy of the material on this Web site for personal, noncommercial use only. The material made available on this Web site is protected by United States copyright laws and is provided solely for the use of instructors in teaching their courses and assessing student learning. Dissemination or sale of any of this material, as a whole or in parts (including on the World Wide Web), is not permitted. All users of these materials and visitors to this Web site are expected to abide by these restrictions. Requests for permission for other uses of this material should be directed to Cold Spring Harbor Laboratory Press, 1 Bungtown Road, Cold Spring Harbor, NY 11724 or submitted via our World Wide Web Site at http://www.cshlpress.com/. |