Properties of the Coalescent Process
If we trace back the ancestry of two genes, there is a chance of 1/2Ne per generation that they will coalesce. Thus, the time until coalescence follows an exponential distribution with mean T2 = 2Ne generations (Table 28.1). If there are k genes, then there are k(k – 1)/2 pairs of lineages which could coalesce (three possible pairs with k = 3, six pairs with k = 4, and so on). Thus, the time until the first coalescence follows an exponential distribution with mean Tk = 4Ne/(k(k – 1)). We can find the expected time TMRCA until the k lineages have coalesced in the most recent common ancestor (MRCA) by adding up the expected time to go from k down to k – 1, from k – 1 down to k – 2, and so on:
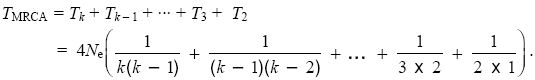
Now, 1/(k(k – 1)) = (1/(k – 1) – 1/k). This leads to a much simpler formula:
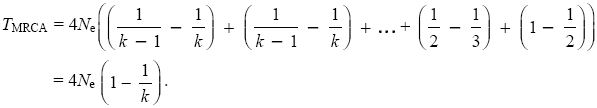
Even with very many lineages (i.e., when k is large), the average time to go down to two lineages is 2Ne, followed by another 2Ne generations to go down to the most recent common ancestor. This is because coalescence is extremely rapid at first, but it then slows down (see Fig. 15.8A).
Other properties of the coalescent process can be found in the same way. For example, the total length of the genealogy (i.e., the total time spanned by all the lineages leading down from the common ancestor to the sample of k genes) is twice the time during which there are two lineages, plus three times the time during which there are three, and so on. The average length is therefore
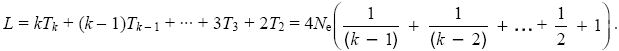
This is close to 4Neloge(1.78k). This result is important, because it determines the average number of mutations that will occur somewhere in the genealogy and will therefore appear as polymorphisms or segregating sites.
|

