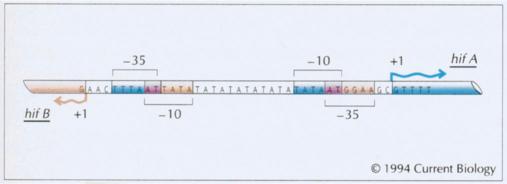

Figure WN23.3. Nucleotide sequence of the intergenic region between the H. influenzae piliation genes hifA and hifB. The positions of the transcriptional start points (+1) and putative promoter elements (–35 and –10) are indicated (in blue for hifA and red for hifB). The loss or gain of TA repeats, ten of which are shown, alters the spacing between the –10 and –35 consensus sequences, which are thought to be binding sites for RNA polymerase. This alters the efficiency with which RNA polymerase binds, thus affecting transcription. With ten TA repeats, the spacing between the –10 and –35 elements is 16 nucleotides, which is predicted to allow efficient binding of RNA polymerase, whereas with nine or eleven repeats, transcriptional efficiency is predicted to be reduced. These predictions correlate with the finding that fimbria expression is maximal with 10 TA repeats, reduced with 11 TA repeats, and absent with 9 TA repeats (van Ham et al. 1993). (Reproduced from Fig. 2 of Moxon et al. 1994.)
| © 2007-2010 by Cold Spring Harbor Laboratory Press. All rights reserved. |
| The reproduction, modification, storage in a retrieval system, or retransmission, in any form or by any means, electronic, mechanical, or otherwise, for reasons other than personal, noncommercial use is strictly prohibited without prior written permission. You are authorized to download one copy of the material on this Web site for personal, noncommercial use only. The material made available on this Web site is protected by United States copyright laws and is provided solely for the use of instructors in teaching their courses and assessing student learning. Dissemination or sale of any of this material, as a whole or in parts (including on the World Wide Web), is not permitted. All users of these materials and visitors to this Web site are expected to abide by these restrictions. Requests for permission for other uses of this material should be directed to Cold Spring Harbor Laboratory Press, 1 Bungtown Road, Cold Spring Harbor, NY 11724 or submitted via our World Wide Web Site at http://www.cshlpress.com/. |