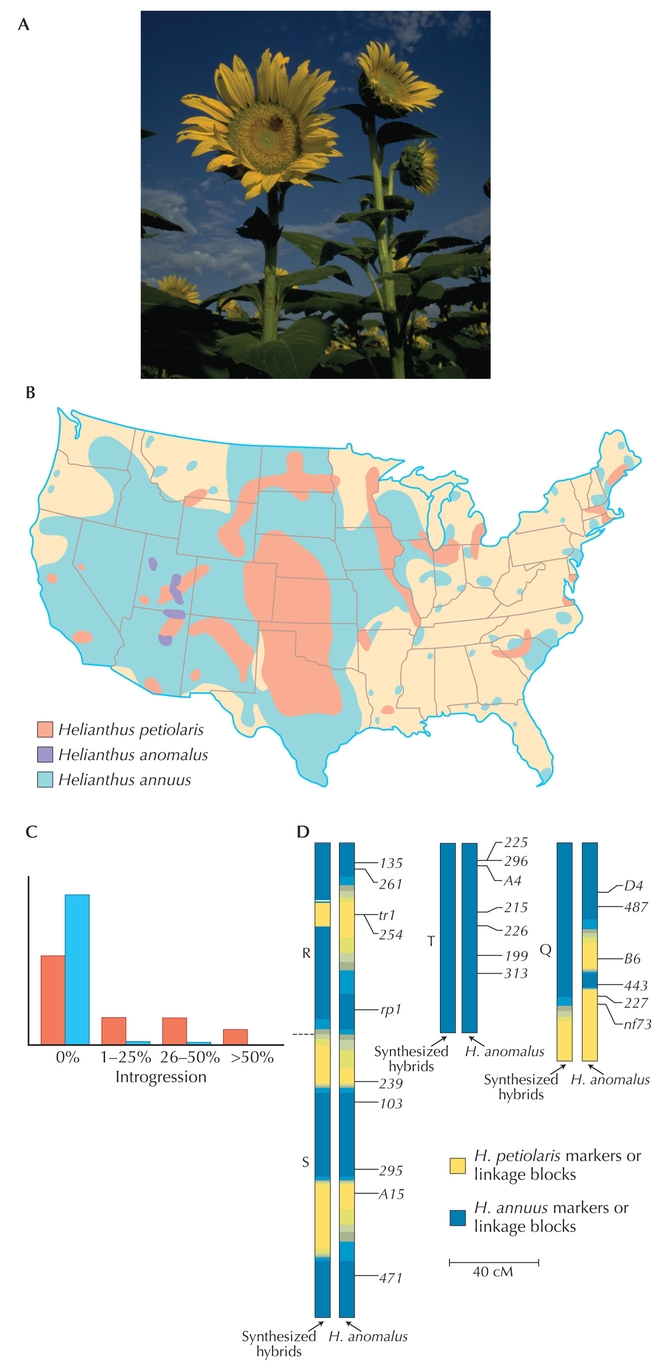

Figure WN22.4. Hybrid speciation in Helianthus. (A) The F1 hybrid, H. anomalus, derived from the parent species, H. annuus and H. petiolaris. (B) The ranges of H. annuus and H. petiolaris, together with that of the hybrid species H. anomalus. (C) The distribution across markers of the proportion of H. petiolaris alleles seen in experimental hybrids. There were three generations of crossing within the hybrid population, followed by two generations of backcrossing to H. annuus. Therefore, in the absence of selection, one expects 1/8 of the genes to derive from H. petiolaris, with a distribution concentrated in the 1–25% class. In regions of genome with the same gene order in H. petiolaris and H. annuus (red ), most markers fail to introgress, but some introgress more than expected. In regions of genome that differ in gene order as a result of chromosome rearrangements, there is almost no introgression (blue). (D) Patterns of introgression along the genomes are similar between experimental hybrids and the natural hybrid species, H. anomalus. Three of the 17 H. anomalus chromosomes are shown. The letters to the left (R, S, T, Q) indicate homology of these chromosomes to regions of the parental genomes. (The leftmost chromosome is rearranged, and combines linkage blocks R and S.) Arrows to the right indicate the genetic markers. The shading indicates the likelihood that the regions derived from H. annuus (blue) or H. petiolaris (yellow). (A, Courtesy USDA; B, redrawn from Rogers et al. 1982; C, data from Table 1 in Rieseberg et al. 1995a; D, redrawn from Fig. 3 in Rieseberg and Noyes 1998.)
| © 2007-2010 by Cold Spring Harbor Laboratory Press. All rights reserved. |
| The reproduction, modification, storage in a retrieval system, or retransmission, in any form or by any means, electronic, mechanical, or otherwise, for reasons other than personal, noncommercial use is strictly prohibited without prior written permission. You are authorized to download one copy of the material on this Web site for personal, noncommercial use only. The material made available on this Web site is protected by United States copyright laws and is provided solely for the use of instructors in teaching their courses and assessing student learning. Dissemination or sale of any of this material, as a whole or in parts (including on the World Wide Web), is not permitted. All users of these materials and visitors to this Web site are expected to abide by these restrictions. Requests for permission for other uses of this material should be directed to Cold Spring Harbor Laboratory Press, 1 Bungtown Road, Cold Spring Harbor, NY 11724 or submitted via our World Wide Web Site at http://www.cshlpress.com/. |