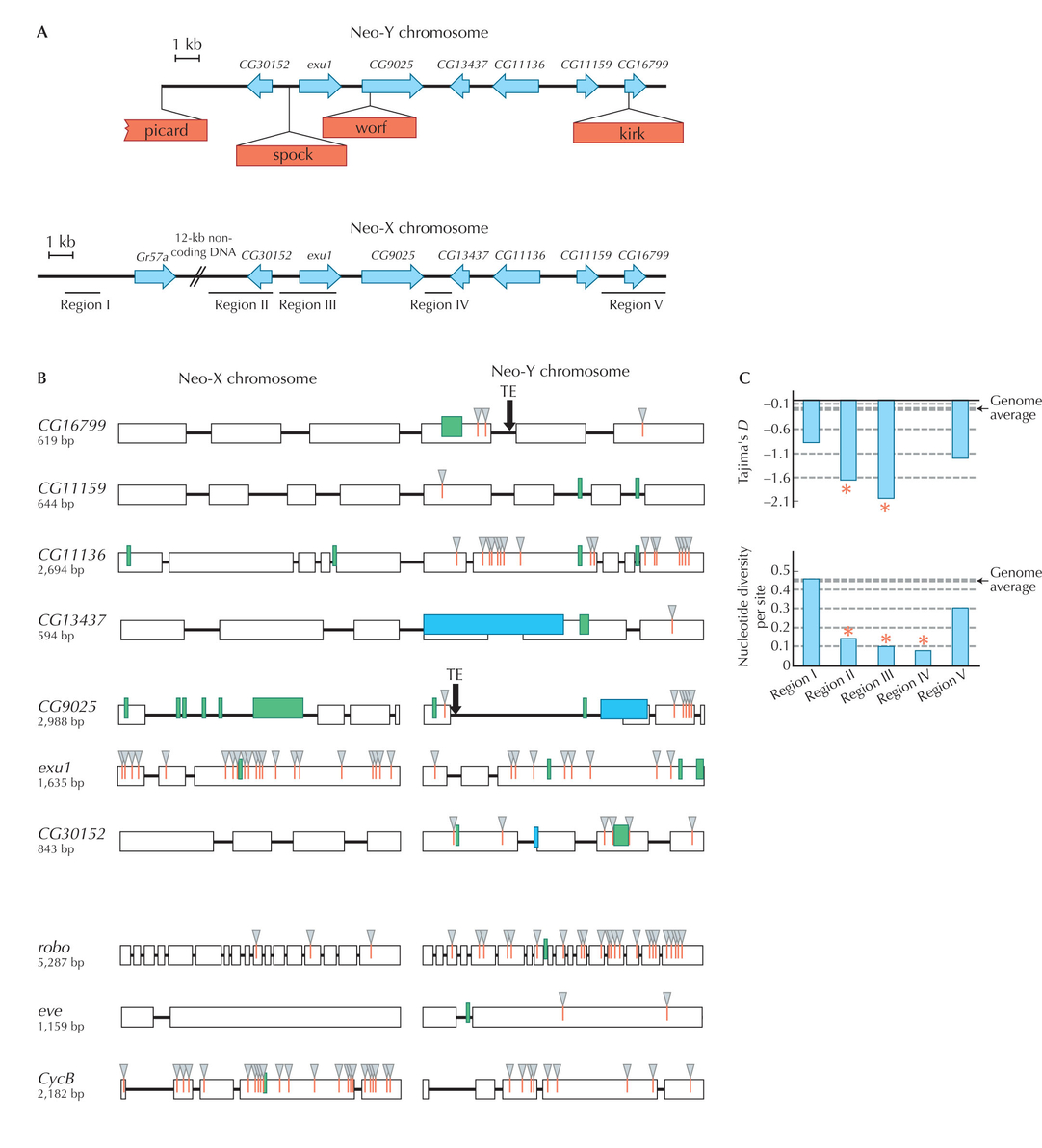

Figure WN23.10. Degeneration of a newly formed Y chromosome in Drosophila miranda. (A) The arrangement of seven genes in an approximately 40-kb region of the neo-X and neo-Y. Note that four transposable elements have fixed in the neo-Y. (B) Divergence in the lineages leading to the neo-X and to the neo-Y were inferred by using D. pseudoobscura as an outgroup. (Gray triangles) Amino acid substitutions; (green boxes) in-frame insertions/deletions; (blue boxes) frameshift mutations. Most genes show an excess of amino acid changes on the neo-Y, indicating degeneration. However, exu1 and cycB show faster amino acid divergence on the neo-X; other evidence shows that this is likely to be due to positive selection. (C) Levels of Tajima’s D statistic and nucleotide diversity, π, across five regions of the neo-Y (see A). Negative Tajima’s D indicates an excess of rare variants; this, together with reduced diversity, is consistent with positive selection at exu1. Asterisk indicates significant deviations from neutrality at the 5% level (Bachtrog 2003). (Redrawn from Figs. 1a, 2, and 1b of Bachtrog 2003.)
| © 2007-2010 by Cold Spring Harbor Laboratory Press. All rights reserved. |
| The reproduction, modification, storage in a retrieval system, or retransmission, in any form or by any means, electronic, mechanical, or otherwise, for reasons other than personal, noncommercial use is strictly prohibited without prior written permission. You are authorized to download one copy of the material on this Web site for personal, noncommercial use only. The material made available on this Web site is protected by United States copyright laws and is provided solely for the use of instructors in teaching their courses and assessing student learning. Dissemination or sale of any of this material, as a whole or in parts (including on the World Wide Web), is not permitted. All users of these materials and visitors to this Web site are expected to abide by these restrictions. Requests for permission for other uses of this material should be directed to Cold Spring Harbor Laboratory Press, 1 Bungtown Road, Cold Spring Harbor, NY 11724 or submitted via our World Wide Web Site at http://www.cshlpress.com/. |