Glossary
This listing includes the full Glossary from the book and additional terms from the online chapters.
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
absolute fitness: See fitness.
acritarch: An organic-walled microfossil, found in ancient rocks, that is interpreted as the reproductive cyst of a eukaryote.
acrocentric: A chromosome with the centromere close to one end.
adaptation: A trait that functions to increase fitness and that evolved for that function.
adaptationist program: An approach to understanding evolution that assumes that traits are adaptations.
adaptive dynamics: A method for finding which phenotypes will invade a population; related to game theory. Alleles that differ slightly from the resident alleles are introduced into a population, and may invade it; if they do, they will either displace the allele, or may coexist with it, in a balanced polymorphism.
adaptive landscape: See fitness landscape.
adaptive peak: A local maximum in the fitness landscape.
adaptive radiation: Diversification of a single lineage into species that exploit diverse ecological niches.
additive genetic variance: The total variance, VA, contributed by the additive effects of each gene (in other words, the variance of breeding values). The response to selection on a quantitative trait is proportional to VA.
additive genetic variance in fitness: The additive genetic variance, VW, of fitness. Fisher’s Fundamental Theorem states that the increase in mean fitness is proportional to this quantity.
additive model: A model in which a quantitative trait is the sum of effects of all the genes involved and of a random environment.
additive tree: A phylogenetic tree in which the branch lengths are proportional to the evolutionary distance between nodes; also known as a phylogram.
ADH: The enzyme alcohol dehydrogenase.
aging: A decrease in survival or reproduction with age. It is equivalent to senescence.
algorithm: A logical process by which a problem can be solved.
align, aligned, alignment: To line up different DNA, RNA, or protein sequences. In most cases alignments include gaps where one molecule has an insertion or deletion relative to others. For phylogenetic analysis, each position (i.e., column) in the alignment should include homologous residues (bases or amino acids) from the different molecules.
allele: A particular form of a gene.
allele frequency: The frequency of a particular allele in a population.
allelic: Two variants are allelic if they are alleles at the same genetic locus.
allometry: Change in proportions with body size. For example, the size of a stag’s antlers relative to its body size increases with body size.
allopatric speciation: The formation of reproductively isolated species due to the divergence of populations that are geographically isolated from each other.
allopatry: The complete separation of populations by geographic barriers.
allopolyploid: A polyploid in which the multiple genomes are derived from different populations or species.
allosteric interaction: See allostery.
allostery: A change in the shape of a protein or ribozyme due to binding of a molecule at one site, which then changes activity at a distant site.
allozyme: One of several variant forms of an enzyme coded by alternative alleles at a single genetic locus.
α-helix: Common structural motif of proteins in which a linear sequence of amino acids folds into a right-hand helix stabilized by internal hydrogen bonding between backbone atoms.
α-proteobacteria: A major class of bacteria that includes many photosynthetic species, many pathogens (e.g., Rickettsias), and many mutualistic symbionts, including the ancestors of mitochondria.
alternative splicing: The process by which the initial RNA made from a single gene can be spliced into different mature messenger RNAs, which in turn produce different proteins.
altruistic: A gene, trait, or behavior that reduces the fitness of its bearer but increases the fitness of other individuals.
alveolate: Member of Alveolata, one of the major kingdoms of eukaryotes. It includes apicomplexans, dinoflagellates, and ciliates.
amelioration: The process in which DNA that has been acquired by lateral gene transfer changes in composition (e.g., G + C content and codon usage) to resemble the genome in which it resides.
AMH: See anatomically modern human.
amniocentesis: Removal of amniotic fluid that surrounds the embryo. The fluid contains fetal cells that can be used for prenatal diagnosis.
amoeba: A single-celled eukaryote that has no fixed shape. This phenotype is found in many eukaryotic lineages.
Amoebozoa: One of the major kingdoms of eukaryotes. Most species are heterotrophic. It includes some species that do not have mitochondria.
amphipathic: A molecule that has both hydrophobic and hydrophilic components (e.g., the phospholipids that make up membranes).
analogy: Similarity that is not due to homology (i.e., common ancestry).
analysis of variance: The separation of the variance into a sum of components, a widely used statistical technique that is the basis for quantitative genetics.
anatomically modern human (AMH): A member of the human lineage that is recognized by anthropologists as having essentially the same anatomy as present-day Homo sapiens.
ancestral characteristic: A trait found in both an organism being studied and the common ancestor of a group to which the study organism belongs.
aneuploid: A cell or organism having an abnormal set of chromosomes.
angiosperm: Flowering plant.
anisogamy: Differentiation of gametes into two (or more) sizes.
annelids: Member of Annelida, a phylum within the Lophotrochozoa. This group of segmented worms includes earthworms, leeches, and bristle worms.
antagonistic pleiotropy: Describes alleles that increase one fitness component but decrease another. It is also a term for theories of aging that involve such alleles.
anther: The male reproductive organ where pollen is produced in a flowering plant.
antibody: A protein that binds to a specific antigen.
antigen: A chemical that triggers an immune response by binding to a specific antibody.
antimutator: An allele or genotype that reduces mutation rates.
antisense oligonucleotide: A short synthetic nucleic acid sequence that is complementary to an mRNA sequence. Through a variety of mechanisms, antisense oligonucleotides can be used to inactivate gene function by interfering with the ability of an mRNA to produce its corresponding protein product.
apicomplexan: Member of Apicomplexa, a phylum of eukaryotes, in the alveolate kingdom. Many species in this phylum are parasites (e.g., Plasmodium falciparum, the causative agent of malaria).
apicoplast: An organelle found in apicomplexan species that is involved in metabolic processes. It is derived from photosynthetic chloroplasts even though it is not now involved in photosynthesis.
archaea: One of the three domains of life. These are species that do not have nuclei and thus were originally grouped with bacteria into the “prokaryotes.” They were identified as a separate domain by analysis of rRNA sequences.
archaic human: A hominin that does not have the modern anatomy of Homo sapiens. Contrast with anatomically modern human.
argument from design: The argument that the order seen in the living world implies that it was created by a divine power.
arthropod: Member of Arthropoda, a major phylum within the ecdysozoa. This group, which is characterized by jointed legs, includes the insects, centipedes, millipedes, spiders, and crustaceans.
association study: A survey of associations between genetic markers and quantitative traits (including human disease) that aims to locate the QTLs that cause trait variation.
ATP: Adenosine triphosphate.
autocatalytic network: A chemical system that outputs a chemical that is a catalyst for the original reaction or that leads to other reactions that eventually output a catalyst for the original reaction.
autoinducer: A chemical used in quorum sensing that is secreted by cells and then used to quantify cell density.
autoinduction: The induction of a regulatory cascade in quorum sensing. It is triggered in response to the crossing of a threshold concentration of autoinducer in the environment.
automatic selection: The increase in frequency of an allele that increases the rate of selfing. It is closely related to the twofold cost of genome dilution.
autopolyploid: A polyploid that carries multiple genomes derived from within the same population.
autosome: A chromosome that is inherited in the usual Mendelian way, in contrast to sex chromosomes and mtDNA.
average effect: The effect of an allele on a quantitative trait as estimated by regression. For a random-mating population that is in linkage equilibrium, this is equivalent to the average excess.
average excess: The difference between the average trait value of individuals who carry a particular allele and the average of the population. For a random-mating population that is in linkage equilibrium, this is equivalent to the average effect.
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
B chromosome: An extra chromosome that is not required for normal function in either sex and is present in only some individuals.
backcross: A cross between a hybrid individual and one of the parental genotypes.
background selection: The reduction in genetic diversity caused by selection against deleterious alleles at linked loci.
bacteria: One of the three domains of life. These are species that do not have nuclei and thus were originally grouped with archaea into the “prokaryotes.”
bacteriocin: A toxin produced by a bacterium that kills its competitors.
bacteriophage: A virus that infects bacteria. The term is commonly used to refer to viruses that infect archaea as well.
balanced polymorphism: A stable polymorphism maintained by balancing selection.
balancer chromosome: A Drosophila chromosome that carries multiple inversions, recessive lethals, and dominant markers. Such chromosomes cannot recombine and cannot become homozygous and so can be used to keep individual wild-type chromosomes intact.
balance view: The view that genetic variation is mostly maintained by balancing selection. This contrasts with the classical view that most variation is due to deleterious mutations.
balancing selection: Selection that maintains polymorphism.
Batesian mimicry: A palatable mimic evolves to resemble a distasteful model species and thereby suffers less predation.
benthic: Living at the bottom of a body of water.
β: See selection gradient.
β-sheet: Common structural motif of proteins in which linear amino acid sequences (“strands”) located in different regions of the polypeptide chain align adjacent to each other and are stabilized by hydrogen bonding between backbone atoms located in different strands.
bilaterian: Member of the group that includes the majority of animal phyla and includes all animals with bilateral (left/right) symmetry.
binomial distribution: If n genes are sampled from a population in which the P allele has frequency p, then the chance of finding j P alleles is
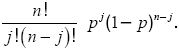
binomial nomenclature: Naming scheme for species in which there is a genus name and a species name.
biological species concept (BSC): Definition of species as groups of individuals that can successfully interbreed with each other in nature but that are reproductively isolated from other such groups.
biometry: The application of statistical methods to biology.
body plan: An organism’s overall morphology that is created by the reproducible spatial positioning of differentiated cell types.
bootstrap: A statistical method for measuring consistency in datasets in which new simulated datasets are generated by sampling with replacement. In phylogenetic inference, bootstrap values represent the percentage of times that a particular clade is seen in trees generated with different simulated datasets.
bottleneck: See population bottleneck.
brachiopod: Member of the phylum Brachiopoda within the lophotrochozoa, composed of a group of marine animals that superficially resemble clams but are only distantly related to the mollusks.
branch: Portion of an evolutionary tree diagram connecting two nodes.
branching process: A process in which individuals produce a random number of offspring, independently of each other.
branch length: The length of a particular branch in an evolutionary tree. In certain types of evolutionary trees, the branch length is used to represent the amount of evolutionary change or time. A scale bar is used to represent the units of measurement.
breeding value: The sum of the average effect of each gene. If an individual is mated at random to others in the population, its offspring will deviate from the population average by one-half the breeding value.
broad-sense heritability (H2): See heritability.
bryozoan: Member of a phylum (Ectoprocta or Bryozoa) of sessile colonial animals, commonly referred to as sea mats or moss animals, that are superficially similar to corals but are instead members of the Lophotrochozoa.
BSC: See biological species concept.
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
C-value: See genome size.
canalization: The buffering of development such that the same form is produced despite genetic and environmental perturbations.
candidate gene: A gene that is thought likely to influence the trait of interest, usually because major mutations at the gene affect the trait.
canonical code: The genetic code that is used almost universally.
capsid: A protein casing that makes up the outside of a virus particle.
case-control study: A form of association study in which “case” individuals with, for example, a disease are compared with “control” individuals without it.
catalysis: The facilitation of a chemical reaction by a molecule that is not itself altered by the reaction.
cDNA: See complementary DNA.
centiMorgan (cM): A distance on the genetic map that corresponds to a 1% recombination rate. See Morgan.
Central Dogma: Information can pass from nucleic acid to protein but not in the opposite direction.
Central Limit Theorem: A theorem that states that the sum of a large number of independent variables tends toward a normal distribution.
centromere: The region of chromosome that attaches to the spindle at mitosis and meiosis.
chaetognath: Any worm of the phylum Chaetognatha, commonly called an arrowworm. This is a transparent marine worm with horizontal lateral and caudal fins and a row of movable curved spines at each side of the mouth.
chaperone: A protein that assists other proteins in achieving a properly folded state.
character state: The particular form that a character trait takes (e.g., a three-chambered vs. four-chambered heart, or an A vs. a T at a particular position in homologous gene sequences).
character state reconstruction: A method used to infer ancestral and derived character states and traits.
character trait: Particular parts or properties of an organism. Used in phylogenetic analysis and to study the evolution of traits over time. Character traits include both discrete (e.g., presence of a heart or globin molecules, type of globins found) and quantitative traits (e.g., four-chambered heart, number of different types of globins).
chemical evolution: Chemical reactions that could have generated complex compounds from simple ones prior to the origin of life.
chemostat: A device that allows populations of microorganisms to be maintained in a steady state.
chert: Very fine grained silica (SiO2) that forms layers or nodules in sequences of sedimentary rocks.
chiasma: The cross-like structure formed by crossing over during meiosis.
chi-square distribution: The distribution of the sum of squares of a number n of normally distributed variables; written as 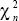 . .
chlorophyte: Member of a phylum of eukaryotes that are all single-celled green algae and closely related to green plants.
chloroplast: A photosynthetic organelle found in many plants, algae, and other microbial eukaryotes. All chloroplasts are evolutionarily derived from cyanobacteria.
choanoflagellate: Member of a phylum of eukaryotes including single-celled flagellated species. This is the sister phylum to animals.
chordate: Member of a major phylum (Chordata) within the deuterostomes, which includes the vertebrates and closely allied invertebrates such as tunicates and amphioxus. All chordates contain a solid rod, called a notochord, along the length of the body during embryogenesis and a dorsal nerve cord and pharyngeal pouches.
chromatid: One of the two copies of a chromosome after it has been replicated.
chromatin: A compact structure of DNA and protein found in eukaryotic nuclei.
chromosomal fusion: See fusion, chromosomal.
chromosomal inversion: See inversion, chromosomal.
ciliate: Member of a phylum of eukaryotes including single-celled species. Most are coated on the outside with cilia, which are used for movement and cellular functions. They are in the kingdom Alveolata.
cis-regulation: Refers to the regulation of when and where a gene is transcribed by DNA sequences that lie to either side (5′ or 3′) of the gene or within the introns of the gene.
clade: A group of species or genes that includes all descendants of an ancestral species or gene. See monophyletic.
cladistics: A method of classification that is based on the order of branching in a phylogenetic tree rather than on phenotypic similarity.
cladogram: A phylogenetic tree in which the only information given is about the relationships among taxa (i.e., the length of the branches is not meaningful).
classical view: The view that genetic variation is mostly due to deleterious mutations. This contrasts with the balance view that most variation is maintained by balancing selection.
cline: A smooth change from place to place across a spatially continuous habitat. The term refers to a spatial gradient in any measurable characteristic—for example, allele frequency or the mean of a quantitative trait.
clonal interference: In an asexual population, different clones, each favored by selection, compete with each other so that only one can succeed.
clone: A set of genetically identical individuals. In genetic engineering, a line of microbes that carry a particular sequence from another species.
cM: See centiMorgan.
CMS: See cytoplasmic male sterility.
cnidarian: Member of a major animal phylum (Cnidaria) that includes corals, sea anemones, hydra, and jellyfish. The group is characterized by the presence of stinging cells called nematocysts.
coalescence: The merging of two genetic lineages into a single common ancestor.
coalescence time: The time back to when two genes share a common ancestor.
coalescent process: A model in which as one moves back in time, each pair of lineages coalesces at a rate 1/2Ne.
coancestry: A measure of the relatedness of two individuals. It is the chance that two randomly chosen genes, one from each individual, are identical by descent.
codon: Three bases that code for a single amino acid.
codon usage: The frequency with which each of the alternative codons that code for an amino acid is used.
codon usage bias: A bias toward use of one of several alternative codons that code for the same amino acid.
coefficient of kinship: See coancestry.
coevolution: The joint evolution of two species, with each responding to selection imposed by the other.
colloid: A substance that contains components in different phases (e.g., minute solid particles within a liquid).
compartmentalization: Subdivision of molecules, cells, or genetic functions into discrete spatial or temporal units. For example, this can refer to the grouping together of a protein into distinct parts of the cell or of networks of genetic interactions into distinct functional units.
competence: The uptake of DNA directly from the environment.
competitive exclusion: Species that use exactly the same resources cannot coexist in a stable equilibrium.
complementary DNA (cDNA): DNA that is complementary to messenger RNA. By producing cDNA using reverse transcription from mRNA, actively expressed genes can be identified.
complementation test: A test for determining whether two mutations are in the same gene (they do not complement) or different genes (they complement).
component of fitness: See fitness component.
component of variance: See variance component.
concerted evolution: The evolution of repeated sequences, which tend to remain homogeneous because of processes such as unequal crossing over and gene conversion.
confidence interval: The range of parameter values that do not deviate significantly from a null hypothesis.
congruency: A measure of how similar are different phylogenetic trees containing the same operational taxonomic units (OTUs).
conjugation: The transfer through a pilus of DNA from one bacterium or archaeon to another.
conservative DNA transposons: A DNA-based transposable element that moves itself to a new place in the genome but does not leave a copy in the original location.
consistency: A measure of whether, given sufficient data, a method will generate a correct answer.
consistency index: A measure of how well the character states for a specific character trait map on a phylogenetic tree. It is calculated as the ratio of the minimum number of evolutionary state changes (aka steps) that could be seen from all possible trees divided by the actual number of changes on the tree being examined.
constitutive: Referes to a gene that is always expressed.
contingency loci: Loci made up of microsatellite repeats (e.g., ATATATAT) in which, when the number of copies of the repeat changes, the phenotype of the cell changes drastically. These are common in the genomes of some bacterial pathogens.
convergence (also convergent evolution): The process by which features with no common ancestry become similar as a result of selection.
correlation coefficient: The most commonly used measure of correlation between two variables (x, y). It was devised by Karl Pearson and is defined as the ratio between the covariance and the square roots of the variances:
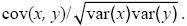
correlation, genetic: See genetic correlation.
cosexual: Producing both male and female gametes. It is equivalent to hermaphrodite.
cost of genome dilution: The disadvantage to a female who reproduces sexually and therefore devotes resources to propagating her mate’s genes.
cost of meiosis: See cost of genome dilution.
cost of natural selection: See substitution load.
covariance: A measure of association between two variables (x, y). It is defined as the expected product of their deviations from the mean:
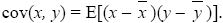
Note that cov(x, x) = var(x).
covariance matrix: An n × n matrix giving the covariances between a set of n variables. The diagonal gives the covariances of each variable with itself (i.e., the variances).
covirus: One of a pair of viruses that have complementary functions and that must coinfect a cell for successful viral transmission.
cristae: Infolded internal membranes, such as those seen in mitochondria and plastids.
crossover: A recombination event within a chromosome at meiosis.
crown group: That part of a clade of living and fossil organisms that includes the last common ancestor of all the living forms and all of its descendants. See stem group.
ctenophore: Member of a major animal phylum of solitary gelatinous marine animals commonly called comb jellies or sea gooseberries (phylum Ctenophora). Members of this group have rows of small combs composed of cilia that are used for locomotion.
cultural evolution: Change in culture (i.e., information passed on by learning and imitation rather than by biological inheritance).
culturing: The growth of a particular microorganism in the laboratory in isolation from other organisms.
cumulative distribution: The probability that a random variable will be less than a given value is called its cumulative distribution.
cyanobacteria: One of the major phyla of bacteria. Most species are photosynthetic, and chloroplasts are derived from this group.
cytology: Study of cells.
cytoplasmic male sterility (CMS): Loss of male function due to a cytoplasmically inherited factor in flowering plants.
cytoskeleton: The system of protein filaments in the cytoplasm of eukaryotic cells that gives the cell its shape and its capacity for directed movement and that participates in the directed transport of molecules within a cell.
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
D: This symbol is used in several ways. See genetic distance and linkage disequilibrium. Dn and Ds are also used as alternative notations for Ka and Ks.
Darwin (d): A unit for the rate of change of morphology. It was introduced by J.B.S. Haldane and is equal to a change by a factor e = 2.718... per million years or 1 part per million per year.
Darwinian Demon: A hypothetical organism with indefinitely high survival and fertility. This is an idea used to emphasize that all real organisms must face constraints on their fitness.
Darwinian medicine: The application of evolutionary principles to medicine.
defective interfering virus (DIV): A virus that has lost some function and that depends on coinfection with intact virus for transmission.
deism: The view that God works through fixed laws of nature; that is, that events occur without supernatural intervention.
deme: A discrete local population.
dendrogram: A phylogenetic tree in which the branch lengths are constrained to all be equidistant from the root; also known as an ultrameric tree.
density-dependent selection: Selection that occurs when relative fitness depends on population density.
derived characteristic: A trait found in an organism that was not present in the common ancestor of a group of organisms being studied.
descent with modification: Darwin’s term for evolution.
deuterostome: Member of one of the two large groups of bilaterian animals including the echinoderms, hemichordates, and chordates. In this group, the initial embryonic opening becomes the anus. See protostome.
developmental constraints: Limits on what kinds of organism can develop.
diapause: A resting stage that allows organisms to survive harsh conditions.
differential equation: An equation that gives the rate of change of a system as a function of its present state.
differentiation: The process by which a cell becomes more and more specialized in its function and morphology through the regulation of gene expression and biochemical activities.
diffusion: Spreading due to the cumulative effect of small random movements.
diffusion approximation: A mathematical approximation that describes diffusion using a differential equation. It describes the spread of chemical concentration or allele frequency through physical space and also the spread of a probability distribution through the space of allele or genotype frequencies.
diffusion equation: An equation that describes how a probability distribution spreads out with time; it applies when random fluctuations are small.
dimer: Two molecules that are bound together. Similarly, trimers and tetramers are clusters of three and four molecules, respectively.
dinoflagellate: Member of a phylum of eukaryotes in the Alveolate kingdom. All are single-celled organisms and many are photosynthetic.
diploid: Carrying two copies of each chromosome.
direct selection: See selection, direct.
directional selection: Selection favoring one allele over another or favoring increased values of a quantitative trait. It is equivalent to positive selection.
discicristate: A kingdom of eukaryotes. It includes the kinetoplastids (e.g., trypanosomes) and euglenoids.
disruptive selection: Selection favoring extreme values of a trait.
distance matrix: A table showing the evolutionary distance between sets of operational taxonomic units (e.g., species or genes).
distyly: A polymorphism with two different arrangements of anther and stigma that promotes outcrossing.
DIV: See defective interfering virus.
divergence: The acquisition of differences after evolutionary separation (e.g., of species).
dizygotic twins: Twins formed from separate zygotes and therefore related in the same way as siblings.
DNA ligation: Chemical connection of two DNA strands. It is used in DNA repair, replication, and other molecular processes.
Dobzhansky–Muller model: A simple model for the evolution of reproductive isolation in which two populations accumulate different alleles. These alleles cause no loss of fitness within the genetic background in which they arose, but derived alleles from different populations are incompatible with each other.
domain of attraction: The domain of attraction of a stable equilibrium is the set of states from which a system will evolve toward that equilibrium.
dominance: If the heterozygote is precisely intermediate between the two homozygotes, there is no dominance. Any deviation from this additive model is described as dominance.
dominance deviation: The difference between the trait value of a genotype and the value expected with no dominance.
dominance theory: An explanation for Haldane’s rule, which assumes that F1 sterility or inviability is caused by recessive alleles.
dominance variance (VD): The variance in a quantitative trait that is caused by dominance. It is defined as the variance of dominance deviations.
dominant: An allele is completely dominant with respect to a certain phenotype if it produces that phenotype when present in either one or two copies.
dominant lethal: See lethal.
dosage compensation: A mechanism that ensures that sex-linked genes are expressed at the appropriate level in both males and females.
drift: See random genetic drift.
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
ecdysozoan: Member of a major subdivision within the protostomes that includes the arthropods, nematodes, and several smaller phyla. Members of this group possess an external covering called a cuticle that molts as the animal grows.
echinoderm: Member of a major phylum within the deuterostomes that includes sea urchins, starfish, crinoids, and sea cucumbers. Although they possess bilateral symmetry initially, adults usually show a fivefold radial symmetry.
ecotype: A genotype adapted to a particular environment.
ectoderm: One of three cell layers found in bilaterian embryos (the other two being the endoderm and mesoderm). The ectoderm goes on to form structures such as the external skin, mouth, and the nervous system.
ectopic recombination: Recombination between repetitive DNA elements found in different regions of the genome (e.g., between transposable elements at different sites). This can lead to chromosome rearrangements and deletions.
EEA: See environment of evolutionary adaptation.
effectively neutral: An allele that alters fitness by less than approximately 1/Ne. Its probability of fixation is approximately the same as if it were absolutely neutral.
effective population size (Ne): The size of the ideal Wright–Fisher population that would give the same rate of random drift as the actual population in question.
efficiency: A measure of the speed at which a method performs.
electrophoresis: A technique in which molecules are pulled through a porous medium by an electric field and so are separated according to their charge and mobility.
endocytosis: The engulfment of material found outside a cell by surrounding it with the cell membrane.
endoderm: One of three cell layers found in bilaterian embryos (the other two being the ectoderm and mesoderm). The endoderm goes on to form structures such as the lining of the digestive system and portions of organs such as the liver, lung, and pancreas.
endomembrane system: Series of intracellular membrane compartments found in eukaryotic cells.
endomitosis: Mitosis with no cell division, which leads to a doubling of ploidy.
endoplasmic reticulum: Eukaryotic membrane compartment involved in translation, folding, and transport of proteins.
endosymbiosis: A symbiosis in which one organism lives within cells of another.
entropy: A quantitative measure of disorder. The Second Law of Thermodynamics states that the entropy of a closed system can never decrease.
environmental deviation: The difference E between the expected trait value of a given genotype and its actual value.
environmental genomics: See metagenomics.
environmental variance (VE): The variance of the environmental deviation, var(E) = VE.
environmental variation: Variation between genetically identical individuals.
environment of evolutionary adaptation (EEA): In evolutionary psychology, the Pleistocene environment in which adaptive human traits evolved.
epigenesis: The development of an organism from a zygote through cell differentiation and formation of morphology. The term also refers to the theory that organisms develop in this way rather than by preformation.
epistasis: Interaction between alleles in their effect on a trait. If a quantitative trait is given by adding up contributions from different loci, then we say that there is no epistasis.
ESS: See evolutionarily stable strategy.
euchromatin: The part of the eukaryote genome that is not condensed and that contains most active genes. It contrasts with heterochromatin.
eugenics: Improvement of the human gene pool through selective breeding.
euglenoid: Member of a class of eukaryotic microorganisms bearing flagella. Most members live in freshwater and many possess chloroplasts.
eukaryote: One of the three domains of life. Its species are characterized by the presence of a nucleus.
eusocial: Fully social organisms in which only one or a few individuals in a colony reproduce.
eutherian mammal: A mammal having a placenta. This includes all mammals except monotremes and marsupials.
evolutionarily stable strategy (ESS): A strategy that cannot be displaced by any alternative. Individuals that play the ESS must be at least as fit as any feasible alternative when the rest of the population is playing the ESS.
evolutionary character state reconstruction: See character state reconstruction.
evolutionary computation: Using evolutionary processes—especially natural selection—to solve computational problems. See genetic programming, genetic algorithm.
evolutionary ecology: Study of the evolutionary consequences of interactions between species and between a species and its environment.
evolutionary game: An interaction between individuals in which the payoff depends on the strategy played by each of them.
evolutionary psychology: A field that applies evolutionary principles to understand universal human traits. It is usually assumed that humans are adapted to their past “environment of evolutionary adaptation.”
Evolutionary Synthesis: The synthesis during the 1930s and 1940s of population genetics with other fields of biology (e.g., paleontology, systematics, and botany).
evolvability: Ability to generate heritable variation that can be exploited by selection.
excavate: Member of a kingdom of eukaryotes. All are single-celled species and none are known to have mitochondria.
exon: A protein-coding region of a protein-coding gene.
exon shuffling: Recombination events that mix exons from two different genes.
expectation: The average value of a function g(x) of a random variable x is called its expectation,
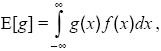
where f(x) is the probability density of x.
expected heterozygosity: See gene diversity.
exploratory system: Systems that shape initially random variation so as to produce a well-coordinated functional outcome.
exponential: Growing at a constant rate r, so that numbers increase with ert.
exponential distribution: A continuous distribution with density λ exp(λx). The distribution of times between events that occur at a consant rate is exponential.
extended phenotype: The phenotype of all the individuals affected by a gene.
extremophile: An organism that thrives in environments that are at the extremes of conditions where life is normally found.
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
falsifiability: A measure of whether the results of a method allow one to determine if assumptions used have been violated.
fitness: The number of offspring left by an individual after one generation. The fitness of an allele is the average fitness of individuals that carry that allele.
fitness component: Traits, such as survival, mating success, and reproduction, that combine to determine fitness.
fitness landscape: Either a graph of fitness as a function of individual genotype or phenotype or of population mean fitness as a function of allele frequencies or trait means.
fitness, mean: The average fitness of a population.
fitness, relative: The fitness divided by the mean fitness or by the fitness of a reference genotype.
fixation probability: See probability of fixation.
fixed (also fix): When all copies of a gene carry the same allele, that allele is said to be fixed.
fluctuation test: An experimental method for measuring mutation rates and for determining whether mutations arise prior to or in response to selection.
founder effect: The loss of genetic variation and the consequent change in genotype frequencies that occurs when a small number of individuals found a new population. It is equivalent to a burst of random genetic drift.
frameshift mutation: An insertion or a deletion mutation that leads to a change in the reading frame in a protein-coding gene.
frequency-dependent selection: Selection that occurs when relative fitness depends on genotype frequencies.
frequency spectrum: The distribution of allele frequencies.
FST: A measure of the genetic diversity between subpopulations relative to the total:
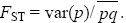
It was devised by Sewall Wright.
Fundamental Theorem (of Natural Selection): The increase in mean fitness due to selection on allele frequencies is equal to the additive genetic variance in fitness divided by the mean fitness:
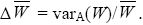
fusion, chromosomal: A mutation in which two chromosomes are connected at their ends. The product is a metacentric chromosome.
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
galls: Structures induced in a plant by a parasite (e.g., a bacterium or an insect) that nurture that parasite.
game: See evolutionary game.
game theory: See evolutionary game.
gametophyte: In plants, the haploid structure that produces gametes. It contrasts with sporophyte.
γ: See selection gradient, quadratic.
Gamma distribution: The sum of a number of independent, exponentially distributed variables (e.g., the time taken for a number of independent events to occur) follows a Gamma distribution.
gas hydrate: Frozen deposits rich in hydrocarbons that occur in the deep ocean basins.
Gaussian distribution: See normal distribution.
gene: A stretch of DNA (or, in some viruses, RNA) sequence that codes for a protein or RNA molecule, together with associated regulatory elements.
genealogical species: A group of individuals for whom the genealogies at all loci in the genome are reciprocally monophyletic. See Web Notes.
genealogy: The tree-like ancestral relationship that connects a set of genes at a single genetic locus.
gene conversion: A meiotic process in which nonreciprocal exchange of genetic information occurs as a result of heteroduplex formation between non-sister chromatids. Thus, heterozygous sites are converted to sites homozygous for one or the other allele.
gene diversity (H): The probability that two randomly chosen genes will carry different alleles. It is equal to the heterozygosity for a randomly mating diploid population with no selection.
gene dosage: The number of copies of a gene within an individual.
gene flow: The movement of genes from place to place. The term usually refers to movement in space but can also refer to movement between microhabitats or to introgression between distinct populations or species.
gene therapy: A treatment in which functional genes are introduced into the patient.
genetic algorithm: An algorithm that applies selection, mutation, and recombination to a population of computer programs in order to solve computational problems.
genetic assimilation: The process in which a phenotype, normally expressed only in a specific environment, through selection shows increased expression in that environment, which may cause the phenotype to be expressed under normal conditions as well.
genetic background: The set of genes with which a gene of interest is associated. Recombination moves genes from one genetic background to another.
genetic code: The code that translates 64 possible triplet codons into amino acids and translation stop signals.
genetic correlation: A correlation between the breeding values for different traits. This may be due to linkage disequilibrium between genes affecting the different traits or to pleiotropy, in which alleles affect both traits.
genetic distance: A measure of the difference in allele frequencies between populations. The most widely used measure of genetic distance is Nei’s D.
genetic drift: See random genetic drift.
genetic engineering: The manipulation of organisms by the artificial introduction of DNA sequence in order to change their characteristics.
genetic load: The loss of mean fitness relative to some ideal fitness:
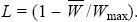
It includes mutation, substitution, recombination, and segregation loads.
genetic locus: See locus.
genetic map: A map of the linear order of genes constructed by measuring the rates of recombination between them.
genetic marker: A polymorphic locus that is used to observe genetic variation but that is not itself of primary interest.
genetic programming: Generating computer programs by selection on a population of variant programs. It is similar to the use of genetic algorithms.
genetic recombination: See recombination.
genetic system: The system immediately responsible for transmission of genetic information.
genome size (C-value): The size of a single haploid genome. It is sometimes expressed as the mass of a single haploid genome measured in picograms (1 pg = 10–12 g) and corresponding to approximately 1000 Mb of DNA.
genomic imprinting: See imprinting, genomic.
genotype: The set of alleles carried by an individual.
genotype frequency: The frequency of a particular genotype in the population.
genotypic value: The average trait value G of individuals with a particular genotype.
genotypic variance (VG): The variance of the genotypic value: var(G)= VG.
geometric mean: An average defined by the nth root of the product of n values:
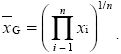
germ line: In a multicellular organism, the lineage of cells that will generate gametes via meiosis. It contrasts with soma.
glaucocystophyte: Member of a phylum of eukaryotes that contains single-celled photosynthetic species.
good genes: Genes that increase fitness for reasons that are not due to sexual selection on a signal trait. Such components of fitness are most conveniently thought of as viability, but fertility and mating success could also be included.
gracile: Graceful, slender, and delicate. The term is used to describe the large rear teeth of certain Australopithecenes. It contrasts with robust.
Gram stain: A stain that specifically detects the type of cell wall and membrane structure found in some bacterial species. These are known as “gram-positive” species.
great plate count anomaly: A phenomenon in which the number of cells from natural environments that can be grown in culture is much less than what can be seen through a microscope. This was one of the first indications of the difficulties in culturing many of the microbes found in the environment.
green beard: An allele that causes a hypothetical trait (a “green beard”) will tend to increase if individuals with “green beards” tend to help others that carry the same trait. This thought experiment shows that kin selection need not depend on a family relationship.
group selection: Selection of traits that increase survival and proliferation of groups of individuals.
gynodioecious: A population that contains both females and hermaphrodites.
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
H: See gene diversity.
H2: See heritability.
h2: See heritability.
Haldane’s rule: When in the offspring of two different animal taxa, one sex is absent, rare, or sterile, that sex is the sex with heterozygous sex chromosomes. Defined by J.B.S. Haldane in 1922.
halophilic: Describes an organism with a preference for growth in high-salt environments.
haltere: Sense organs found in Diptera on the second thoracic segment, evolutionarily derived by modification of the wings. They are used to help balance during flight.
Hamilton’s rule: An allele that increases the fitness of its neighbors by B but reduces the fitness of its carrier by C will increase if rB > C, where r is a measure of the genetic similarity of neighbors to the focal individual.
handicap: A trait that signals a male’s genetic quality. Its association with good genes is maintained because it is less costly to males of higher quality.
haplodiploid: A system of sex determination in which fertilized eggs develop as diploid females and unfertilized eggs develop as haploid males.
haploid: Carrying one copy of each chromosome.
haplotype: A particular combination of alleles in a haploid—that is, a haploid genotype.
hard selection: Selection where numbers produced by a patch are directly proportional to the relative survival of the individuals of that patch, see Web Notes.
Hardy–Weinberg proportions: The frequencies of diploid genotypes produced after random mating. In the simplest case of two alleles at frequencies q + p = 1, the three genotypes QQ, PQ, PP are at frequencies q 2, 2pq, p 2.
harmonic mean: An average defined by
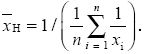
It gives greatest weight to small values. The effective population size is the harmonic mean of the population sizes in each generation.
hemichordate: Member of a diverse phylum of marine animals including the acorn worms and pterobranchs (phylum Hemichordata).
hemimetabolous: Developing directly through a series of nymphal stages with a similar morphology to the adult (e.g., as in grasshoppers and bugs). It contrasts with holometabolous.
heritability: The fraction of phenotypic variance that is inherited. The broad-sense heritability refers to the total genotypic variance (H2= VG/VP), whereas the narrow-sense heritability refers to the additive genetic variance (h2= VA/VP).
hermaphrodite: An individual that produces both male and female gametes.
heterochrony: The change in the relative timing or duration of events during development achieved by altering the relative onset or ending of particular developmental processes.
heterogametic: The sex that carries distinct sex chromosomes. For example, in mammals, males are the heterogametic sex.
heterokont: One of the kingdoms of eukaryotes. It includes a diverse collection of single-celled species including diatoms.
heterosis: The increase in fitness seen in a cross between different populations.
heterostyly: A polymorphism for distinct arrangements of anther and stigma.
heterozygosity: The proportion of heterozygotes in a population.
heterozygote: A diploid individual that carries two different alleles at a locus.
heterozygote advantage: See overdominance.
hierarchical classification: A system of classifying organisms in a nested series of levels—domain, kingdom, phylum, class, order, family, genus, species.
Hill–Robertson effect: The interference between selection at linked loci. It was first analyzed by Hill and Robertson in 1966.
hitchhiking: The increase in a neutral allele that happens to be associated with a selectively favorable allele at another locus. It is sometimes used more broadly to refer to any kind of indirect selection.
HIV: See human immunodeficiency virus.
HLA: Human leukocyte antigen. See major histocompatibility complex.
Holliday junction: The cross-like structure formed by crossing over between two DNA double helices.
holometabolous: Metamorphosis through a pupal stage (as in flies, butterflies, and beetles). It contrasts with hemimetabolous.
homeobox: A sequence, approximately 180 nucleotides long, that is translated into a DNA-binding domain called the homeodomain. This sequence is found in many transcription factors that play a role in pattern formation and cell differentiation.
homeodomain: A sequence, approximately 60 amino acids long, that is encoded by a homeobox DNA sequence. This protein motif forms a DNA-binding domain that is found in many transcription factors that play a role in pattern formation and cell differentiation.
homeotic: Describes a class of mutations that transforms one part of an organism into another part. For example, a particular homeotic mutation in Drosophila transforms the antennae into legs.
hominid: Member of the great apes (family Hominidae), which now include human, gorilla, orangutan, and chimpanzee.
hominin: All taxa closer to humans than to chimpanzee. Apart from ourselves, all these taxa are now extinct.
homologous recombination: The process by which two pieces of DNA, identical or nearly identical in sequence (e.g., two copies of a chromosome), align and exchange a portion of DNA.
homology: A similarity attributable to its derivation from the same ancestral feature. In genetic studies, it refers to genes that are present in the same locus in the genome. In phenotypic studies, it refers to character traits or states that are present in a set of species and their common ancestor.
homoplasy: A similarity of traits that is not due to homology but instead to convergence or parallel evolution.
homozygote: A diploid individual that carries the same allele at a genetic locus.
homunculus: A “little man” that was supposedly introduced into a fertilized egg by the sperm and that guided its development. See preformation.
horizontal transmission: Transmission of genetic information between different individuals other than from parent to offspring.
host races: Genetically distinct populations that specialize on different hosts.
hot spots, recombination: See recombination hot spots.
human immundeficiency virus (HIV): The virus responsible for AIDS.
hybrid rescue allele: An allele that alleviates hybrid sterility or inviability.
hybrid zone: A narrow region in which genetically distinct populations meet, mate, and hybridize.
hydrogenosome: An organelle of some eukaryotes that produces hydrogen gas and ATP. It is possibly derived from mitochondria.
hydrophilic: A molecule or portion of a molecule that readily dissolves in water via the formation of hydrogen bonds.
hydrophobic: A molecule or portion of a molecule that does not readily dissolve in water.
hydrophobic core: A portion of a protein that avoids dissolution in water and is composed of a set of hydrophobic amino acids.
hypercycle: Cooperation between a set of replicating molecules (A, B, ..., Z), in which A aids replication of B, B aids C, ..., and Z aids A. It was proposed as a way for early life to replicate genetic information despite high mutation rates.
hyperthermophile: An organism that thrives at temperatures above 80°C.
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
IBD: See identity by descent.
identical twins: Twins formed from a single zygote and therefore genetically identical. See monozygotic twins.
identity by descent (IBD): Genes that are inherited from the same gene in an ancestral population are identical by descent.
immune memory: The increased response of the immune system to an antigen that had been encountered before. This is the basis of vaccination.
immunoglobulin: A family of proteins involved in the immune system. It includes antibodies.
immunological distance: A measurement of phylogenetic relationship based on differences in antibody–antigen binding affinity between species. The magnitude of the difference is used as a rough evolutionary clock.
imprinting, genomic: A phenomenon in which the phenotype of a heterozygote depends on which allele came from the father and which from the mother.
inbred: Produced by mating between relatives.
inbred line: A population produced by continued self-fertilization or mating between close relatives. After several generations of inbreeding, an inbred line becomes genetically homogeneous.
inbreeding: Mating between relatives.
inbreeding coefficient: The chance that two homologous genes in a diploid individual are identical by descent.
inbreeding depression: Reduced vigor of inbred individuals.
inclusive fitness: A measure of individual fitness that includes the effects of that individual on its neighbors’ fitness, weighed by their relatedness, and discounts the effects of neighbors on the individual’s own fitness.
incomplete dominance: A condition in which heterozygotes express a trait that is distinguishable from both homozygotes.
indel: An insertion or a deletion mutation involving a small number of bases.
indirect selection: Selection on a trait that arises from its association with other traits rather than because of its direct causal effect on fitness. The term may also refer to selection that arises from the effects of a gene in one individual on the fitness of other individuals.
induction: Increase in gene expression in response to a regulatory signal.
infinite-alleles model: A model that assumes that every mutation generates a new allele.
infinitesimal model: A model that assumes that quantitative trait variation is caused by a very large number of loci, with infinitesimally small additive effects. Selection alters the mean, but the genetic variance remains constant. It is equivalent to assuming that offspring are normally distributed around the mid-parental value, with constant variance.
infinite-sites model: A model that assumes a very large number of sites, each with a very low mutation rate, so that every mutation is unique.
informational genes: Genes involved in core “informational” processes including DNA replication and repair, transcription, and translation. They are thought to be less prone to lateral gene transfer than operational genes.
ingroup: A set of operational taxonomic units for which the phylogenetic relationships are being determined relative to an outgroup.
inheritance of acquired characteristics: Transmission of characteristics acquired during an organism’s lifetime to its offspring. This view is associated with Lamarck, but it was widely held until the establishment of genetics.
innovation: A change to a preexisting feature.
insertion sequence (IS): A class of transposable elements found in bacteria and archaea.
in situ hybridization: A labeled DNA or RNA probe is hybridized to a tissue section or whole embryo and viewed under the microscope to determine when and where a specific mRNA is expressed. Alternatively, the probe is hybridized to a chromosome spread and viewed under the microscope to determine the location of a specific genomic sequence.
intelligent design: The argument that organisms are irreducibly complex and so must have been created by an intelligent designer. It is a version of the argument from design.
interaction variance: The variance in a quantitative trait, VI, caused by epistatic interactions between loci. It is the difference between the genotypic and the additive plus dominance variance and is VI = VG – VA – VD.
intercalating agent: A chemical that resembles DNA bases and can insert into DNA backbones during replication, leading to insertion or deletion mistakes in replication.
introgression: Movement of genes from one genetic background to another, as a result of hybridization between individuals from distinct populations.
intron: A noncoding sequence that interrupts the coding sequence.
invention: A fundamentally new feature of an organism.
inversion, chromosomal: A mutation in which a section of DNA sequence in the offspring is inverted relative to its orientation in the parent.
irreducibly complex: A system that cannot function if any one of its components is missing.
IS: See insertion sequence.
island model: The simplest model of population structure. A fraction m of the genes in a deme comes from outside. Immigration may be from the mainland population or from other demes.
isogamous: Producing a single kind of gamete.
isolation by distance: Divergence between allele frequencies in different places within a spatially continuous population. The term usually refers to divergence caused by random genetic drift.
isotope: Forms of an element that differ in atomic mass.
isozymes: Enzymes with different amino acid sequences that catalyze the same reaction. This includes variants coded by different genetic loci as well as allozymes, which are coded by homologous genes at the same locus.
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
jackknifing: A statistical method in which new data sets are generated by resampling an original data set without replacement.
junk DNA: Sequences that accumulate by mutation and that are neutral or deleterious.
just-so stories: Untestable explanations for adaptations.
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
Ka: The rate of nonsynonymous substitutions, which alter amino acid sequence. This is sometimes denoted Dn.
kerogen: A class of organic compounds found in some sedimentary rocks and thought to be derived from organic molecules from living organisms.
kinase: An enzyme that adds phosphate groups onto other molecules.
kin discrimination: The ability to distinguish between related and unrelated individuals.
kinetoplast: An independently replicating organelle lying near the base of the flagellum in kinetoplastids.
kinetoplastid: Member of a phylum of eukaryotes in the Excavata kingdom characterized by the presence of a kinetoplast organelle.
kinorhynch: Member of a phylum of tiny spiny animals (phylum Kinorhyncha).
kin selection: A change in the frequency of an allele, caused by the effect of that allele on the fitness of other individuals who carry the allele.
Ks: The rate of synonymous substitutions that do not alter amino acid sequence. It is sometimes denoted Ds.
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
labyrinthulid: Member of a phylum of eukaryotes in the heterokont kingdom.
lagging strand: During DNA replication, the strand that is synthesized in the 3′ to 5′ direction by ligation of short DNA strands synthesized discontinuously in the 5′ to 3′ direction.
Lamarckism: See inheritance of acquired characteristics.
large X effect: The observation that the X chromosome is involved in hybrid incompatibilities more often than would be expected from its size.
last universal common ancestor (LUCA): The common ancestor of all modern life-forms.
lateral gene transfer (LGT): The transmission of DNA from one evolutionary lineage to another. Also known as horizontal gene transfer.
leading eigenvalue: Close to equilibrium, a system can be described by a set of linear equations. These have a set of special solutions that grow exponentially. The rates of growth of these special solutions are called the eigenvalues, and the largest is called the leading eigenvalue. It is this that dominates the behavior of the system near equilibrium.
leading eigenvector: Small deviations {x1, x2, ...} from equilibrium grow exponentially at a rate given by the leading eigenvalue λ and have the form xi = eiexp(λt). The solution {e1, e2, ...} is called the leading eigenvector; it is the direction in which perturbations from equilibrium tend to grow or shrink.
leading strand: During DNA replication, the strand that is synthesized in the 5′ to 3′ direction by continuous polymerization at the growing 3′ tip.
lek: An arena where males gather and are chosen as mates by females.
lethal: A recessive lethal allele kills its bearer when homozygous, whereas a dominant lethal allele kills when present in a single copy.
Levene model: A model of a structured population in which individuals from a single gene pool spend part of their lives competing within small patches.
LGT: See lateral gene transfer.
life history: An organism’s pattern of survival and reproduction.
life-history trait: Traits, such as mortality rate, fertility, or age of reproduction, that are closely associated with fitness.
likelihood: Given a hypothesis, the probability of observing certain data.
likelihood ratio: A ratio of the probability of generating a particular data set given one hypothesis relative to the probability of generating the same data given another hypothesis ((Prob(D|H1)/Prob(D|H2)). This is the value calculated in likelihood-based phylogenetic reconstruction methods.
limit cycle: A system may tend toward an unending cycle rather than a stable equilibrium.
limnetic: Occurring in the deeper open water of lakes or ponds.
LINE: See long interspersed nucleotide element.
lineage sorting: The process by which, following separation of two species, the ancestry of every gene converges to the overall phylogeny of the species. This process takes about 2Ne generations, and incomplete lineage sorting implies discordance between genealogies.
linear equation: An equation of the form y = a + bx, where the variable x does not appear as a power or special function.
linkage: Genes that are carried on the same chromosome are said to be linked.
linkage disequilibrium: Nonrandom associations between alleles at two or more genetic loci.
linkage equilibrium: Absence of linkage disequilibrium, so that haplotype frequencies are equal to the product of allele frequencies.
liposome: A spherical lipid bilayer.
LMC: See local mate competition.
load: See genetic load.
load, mutation: See mutation load.
load, recombination: See recombination load.
load, segregation: See segregation load.
load, substitution: See substitution load.
lobopod: The fossil members of a phylum of caterpillar-like animals. The living forms are terrestrial and commonly referred to as velvet worms. The fossils are marine and were diverse in the Cambrian (phylum Onychophora).
local mate competition (LMC): Competition for mates within a local group (e.g., between fig wasps within a single fig).
locally stable: An equilibrium is locally stable if any sufficiently small perturbation decreases in magnitude. Such an equilibrium is surrounded by a domain of attraction.
locus: A location on the genome. It may refer to a single nucleotide site or to a substantial stretch of DNA sequence.
long-branch attraction: A phenomenon in phylogenetic analyses when taxa or OTUs that lie at the end of long branches are inferred to be closely related, regardless of their true evolutionary relationships.
long interspersed nucleotide element (LINE): A class of transposable element.
lophotrochozoan: Member of a major subdivision within the protostomes that includes the annelids, mollusks, bryozoans, brachiopods, and several additional small phyla.
LUCA: See last universal common ancestor.
lycophyte: Member of a diverse group of early land plants including the lycopods and zosterophylls.
lycopod: Member of a group of plants that includes giant trees in the Carboniferous coal swamp forests and the living club mosses.
lycopsid: See lycopod.
lysogenic: Describes bacteria and archaea that have bacteriophage integrated into their genomes. The bacteriophage may be activated, leading to lysis of the cell.
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
macroevolution: Evolutionary change at or above the species level.
macronucleus: The larger of the two nuclei found in ciliate cells. It functions in ways similar to the somatic cells of animals.
major histocompatibility complex (MHC): A set of closely linked genes in vertebrates that play a key role in the immune response. In humans, it is known as the HLA (human leukocyte antigen) complex.
major transitions: Identified by Maynard Smith and Szathmáry as major changes in the way hereditary information is transmitted. Almost all involve the coming together of previously independent replicators to form more complex entities.
mask: A method for identifying which positions in a multiple sequence alignment to use for phylogenetic reconstruction.
mating types: A polymorphism in which individuals can mate only with a different type. See self-incompatibility.
mean: Usually refers to the arithmetic mean: for n values, z1, ..., zn, 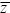 = (Σizi)/n. See also the harmonic mean and the geometric mean. = (Σizi)/n. See also the harmonic mean and the geometric mean.
mean fitness: See fitness, mean.
meiosis: A cellular division process that is involved in sexual reproduction in eukaryotes in which gametes are produced having half the number of copies of each chromosome as the parents.
meiotic drive: Preferential movement of a chromosome during meiosis toward the pole of the cell that will go on to produce gametes.
mesoderm: One of three cell layers found in bilaterian embryos (the other two being the ectoderm and endoderm). The mesoderm goes on to form structures such as the muscles and internal bones and portions of organs such as the kidney and reproductive system.
mesophile: An organism that prefers to live at moderate temperatures.
Mesoproterozoic: Division of time from 1600 to 1000 Mya.
messenger RNA (mRNA): The RNA molecule that is transcribed from the DNA and takes sequence information to the ribosome, where it is translated into protein.
messenger RNA processing: The collective term for the modifications to eukaryotic RNA that are necessary before the RNA can be transported to the cytoplasm for translation.
metagenomics: Large-scale sequencing of DNA isolated directly from environmental samples (e.g., soil, air, and water).
metapopulation: A collection of discrete demes. The first metapopulation models involved extinction and recolonization, but the term is now used more broadly.
metazoa: The group that includes all animal phyla, including sponges, ctenophores, cnidarians, and the bilaterians.
MHC: See major histocompatibility complex.
microarray: An array of short oligonucleotides, bound to a substrate, that can be used to simultaneously measure the concentration of large numbers of different DNA or RNA sequences.
microfilament: Minute fibers composed primarily of actin involved in the structural integrity and movement of eukaryotic cells. Together with microtubules these make up the cytoskeleton.
micronucleus: The smaller of the two nuclei found in ciliate cells. It functions in ways similar to the germ cells of animals.
microRNA: A family of RNA molecules, approximately 22 nucleotides long, that regulates the expression of some eukaryotic genes.
microsatellite: A short array of repeated sequences, each a few base pairs long. Microsatellites tend to be highly polymorphic and so are widely used as genetic markers.
microsporidia: A group of single-celled eukaryotes that were once considered to be their own phylum but are now considered part of the fungal phylum.
microtubule: A major component of the cytoskeleton, composed of the protein tubulin. It is used by eukaryotic cells to regulate their shape and control their movements.
midpoint rooting: A method of rooting a phylogenetic tree in which the root is placed at the center of a branch connecting the pair of operational taxonomic units that are most distant from each other.
migration: Movement from place to place. Here, it is used synonymously with gene flow.
mimicry: An adaptive resemblance between one organism and another. See Batesian mimicry and Müllerian mimicry.
minisatellite: Multiple copies of short sequences, from 9 base pairs up to several hundred base pairs. They are scattered over many sites, with 10–100 repeats, and are highly variable and hence useful as genetic markers.
mismatch distribution: The distribution of numbers of differences between random pairs of sequences sampled from a population.
mismatch repair: The process of repairing simple DNA replication errors such as base misincorporation and small insertions or deletions.
missense mutation: A nucleotide substitution within a protein-coding region of a gene that leads to the replacement of one amino acid by a different amino acid.
mitochondria: The eukaryote organelles responsible for aerobic respiration. Mitochondria are derived from α-proteobacteria.
mitochondrial DNA (mtDNA): The genome contained within mitochondria. Animal mtDNA is widely used as a genetic marker, because it does not recombine.
mitosis: Cellular division process that is involved in asexual reproduction in eukaryotes in which each daughter cell gets a copy of the chromosomes of the parent.
mixed strategy: Where individuals play two or more strategies at random.
modifier: In the strict sense, an allele that has no direct effect on fitness but does have some other effect on the genetic system that may be indirectly selected. More broadly, it refers to an allele that alters the expression of alleles at other loci.
modularity: Subdivision into distinct parts or modules that can function independently. In evolutionary psychology, the term refers to independent mental functions.
molecular clock: The constant rate of accumulation of amino acid or DNA sequence differences.
molecular recognition: The binding of two molecules though noncovalent bonds in which the shape of the molecules plays a key role in the strength of binding.
molecular recombination: The physical cutting and joining of DNA molecules.
mollusks: A major phylum within the lophotrochozoa. This group of animals includes clams, mussels, chitons, octopus, squid, and nudibranchs.
monandrous: Where females mate with a single male.
monophyletic: Describes a set of taxa that all descend from a common ancestral taxon—that is, a group of organisms or genes that share a common ancestor to the exclusion of all other entities.
monosomy: The presence of only one chromosome instead of a pair in a cell’s nucleus.
monozygotic twins: See identical twins.
Morgan: The unit of distance on the genetic map. Over short map distances, the distance between loci in Morgans equals the recombination rate between them. Over long distances, the recombination rate is lower than the map distance because of multiple crossovers.
mRNA: See messenger RNA.
mtDNA: See mitochondrial DNA.
μ: See mutation rate.
Müllerian mimicry: A distasteful species evolves to resemble another distasteful species. Both gain increased protection, because predators learn to avoid the common pattern more quickly.
Muller’s ratchet: The degeneration of an asexual population that arises from the random and irreversible loss of the fittest genotype.
multiple testing: If many significance tests are carried out, then some will reject the null hypothesis just by chance. Thus, significance levels must be reduced when multiple tests are performed.
multiregional model: The hypothesis that different hominins found across the Old World (such as Homo erectus and Homo neanderthalensis) evolved in situ into modern human populations. It contrasts with the out-of-Africa model.
mutation: A heritable change in the genetic material of an organism that does not involve reciprocal recombination.
mutational heritability: The ratio, VM/VE, between the mutational variance and the environmental variance.
mutational variance (VM): The variance in a quantitative trait caused by new mutations in each generation.
mutation load: The loss of mean fitness caused by deleterious mutations:
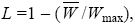
where Wmax is the fitness of the wild-type genotype that carries no mutations.
mutation rate (μ): The rate at which mutations are generated.
mutator: An allele that causes an increased mutation rate.
mutualism: An interaction between species from which all involved gain.
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
NADPH: Nicotine adenine dinucleotide phosphate (in its reduced form). It is used as an energy and redox carrier in all organisms.
narrow-sense heritability (h2): See heritability.
naturalistic fallacy: The mistaken argument that what is justifies what should be—for example, that evolution, or the mechanism of evolution, justifies particular moralities.
natural logarithm: The natural logarithm log(x) is the inverse of the exponential function: log(exp(y)) = y. Sometimes written as loge(x) or ln(x).
natural selection: The process by which genotypes with higher fitness increase in frequency in a population.
natural theology: A theology based on reason and ordinary experience instead of on special or supposedly supernatural revelation.
Ne: See effective population size.
nearest-neighbor exchange: A method for searching phylogenetic tree space whereby a new tree is generated from a starting tree by swapping neighboring branches in the starting tree.
nearly isogenic line (NIL): A line produced by continued crossing of one line back to another, combined with selection. An NIL contains small segments of genome derived from one line, which are enriched for QTLs that influence the selected trait.
neighbor joining: A recursive distance-based phylogenetic reconstruction algorithm that takes as its input a distance matrix and produces an unrooted phylogenetic tree.
neighborhood size: Proportional to the product of the rate of gene flow, σ2, and the effective population size per unit area (i.e., the effective density), ρ, which determines the relative rate of gene flow and random drift in a spatially continuous population. In two dimensions, it is defined as Nb = 4πρσ2.
nematode: Member of a major phylum within the Ecdysozoa of very diverse and abundant worm-like animals, including the round worms and thread worms (phylum Nemata).
Neoproterozoic: Division of time from 1000 Mya to the base of the Cambrian at 542 Mya.
neoteny: Reproduction by juveniles. For example, axolotls reproduce without metamorphosis into adult salamanders.
neutral: Having no effect on fitness.
neutral evolution: Evolving without the influence of natural selection.
neutralist: One who holds that most molecular divergence and variation has negligible effect on fitness, at least at the time when it evolved.
neutral mutation: A mutation that does not affect fitness.
neutral theory: The theory that genetic variation is neutral and is shaped primarily by mutation and random genetic drift.
niche: The set of ecological environments in which a species can survive and reproduce.
NIL: See nearly isogenic line.
node: The point in a phylogenetic tree where one branch splits into two.
nonhomologous gene displacement: A lateral gene transfer event in which a gene that carries out a particular function is replaced by a nonhomologous gene that carries out a similar function.
nonlinear equation: An equation that is not linear.
non-Mendelian inheritance: Inheritance that does not follow Mendelian patterns. (Linkage and sex linkage, which were not known to Mendel, are not included.)
nonsense mutation: A point mutation in a protein-coding region that produces a stop codon, prematurely truncating the protein sequence.
nonsynonymous mutation: A point mutation in a protein-coding region that changes a codon such that it alters the resulting amino acid sequence of the protein.
normal distribution: The bell-shaped curve that describes the distribution of the sum of a large number of independent variables.
nucleolus: Subcompartment within the nucleus that is involved primarily in making ribosome components.
nucleomorph: A highly reduced relic of a nucleus. It is found in many eukaryotic species that have undergone secondary symbioses in which one eukaryote became the endosymbiont of another eukaryote.
nucleotide: A nitrogenous base attached to a ribose or deoxyribose sugar and a phosphate molecule. It is the basic unit of a nucleic acid.
nucleotide diversity (π): The chance that two randomly chosen copies of a nucleotide site will carry different bases. It is similar to gene diversity and applies to single nucleotides.
nucleotide site: A particular nucleotide in the DNA or RNA sequence.
null allele: An allele of a gene in which function has been completely abolished. It is generally considered equivalent to the complete deletion of the gene.
null hypothesis: A hypothesis that is presumed true and against which alternative hypotheses are tested statistically.
null model: See null allele.
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
Occam’s razor: The general principal that if all else is equal, the simplest explanation is best. It is applied when using parsimony methods during phylogenetic reconstruction.
odds ratio: See posterior odds ratio.
Okazaki fragments: Small fragments of DNA made during DNA replication of the lagging strand.
oligonucleotide: A short piece of DNA, no more than about 20 base pairs long.
onychophoran: Member of a phylum of caterpillar-like animals. The living forms are terrestrial and are commonly referred to as velvet worms; the fossils are marine and were diverse in the Cambrian (phylum Onychophora).
open reading frame (ORF): A section of a genome that contains the codons used to make a protein. In bacteria and archaea open reading frames are usually found in contiguous stretches in the genome; in eukaryotes they are frequently interrupted by introns.
operational gene: Genes involved in metabolic and other peripheral processes. They are meant to contrast with informational genes and are thought to be more prone to lateral gene transfer than informational genes.
operational taxonomic unit (OTU): Any entity (e.g., an organism, gene, species, or population) used for phylogenetic study.
operon: A set of adjacent genes whose transcription is regulated as a single unit.
opisthokont: Member of a kingdom of eukaryotes including the metazoa, fungi, and choanoflagellates.
ORF: See open reading frame.
orthogenesis: An inherent tendency for lineages to change in a particular direction.
orthologous: See orthologous genes.
orthologous genes: Genes that are homologous (share a common ancestry) and have diverged from each other due to the separation of the species in which the genes are found (e.g., α-globin from humans and mice). Contrast this with paralogous genes.
OTU: See operational taxonomic unit.
outcrossing: Mating with unrelated individuals.
outgroup: An organism or gene from an evolutionary lineage that separated from those lineages being studied prior to the existence of their common ancestor.
out-of-Africa model: The hypothesis that modern humans evolved recently in Africa and spread from there, replacing archaic hominins. It contrasts with the multiregional model.
overdominance: Describes heterozygotes that have higher trait values (usually higher fitness) than either homozygote.
ovule: The female gamete in a flowering plant. It is analogous to the egg in animals.
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
Paleoproterozoic: Division of time from 2500 to 1600 Mya.
panmictic: Describes a population in which every individual has the same chance of mating with every other: in other words, where there is no population structure.
paracentric inversion: A mutation that involves a chromosomal inversion that does not span the centromere.
parallel evolution: The process by which features that once were different become similar by experiencing the same changes in different evolutionary lineages.
paralogous genes: Genes that are homologous (share a common ancestry) and have diverged from each other after gene duplication events (e.g., α- and β-globins). Contrast this with orthologous genes.
parameters: Quantities such as selection coefficients or recombination rates that determine the behavior of a model but do not evolve with it. Contrast with variables.
parapatric: A geographic distribution in which different types are found in different places and meet only in a narrow zone.
parapatric speciation: The evolution of new species within a spatially extended population in the presence of gene flow.
paraphyletic: Describes a group of organisms or genes that share a common ancestor to the exclusion of all other entities but in which some members of the group are excluded.
parsimony: General approach to evolutionary reconstructions in which the goal is to identify theories (e.g., evolutionary branching patterns) that require the fewest number of evolutionary events (and thus might be considered the simplest).
parthenogenesis: The production of offspring from unfertilized eggs.
pathogenicity islands: Contiguous sections of a pathogen’s genome that contain a disproportionate (relative to number of base pairs) number of the factors that cause pathogenicity.
patrilineal: Inherited from the father (e.g., the Y chromosome in mammals).
payoff: In an evolutionary context, the increase in fitness due to a contest.
payoff matrix: A matrix showing the payoff that is won by each possible strategy when played against each other strategy.
PCR: See polymerase chain reaction.
pedigree: The family relationships between individuals in a sexual population.
pericentric inversion: A mutation involving a chromosomal inversion that spans the centromere.
peroxisome: A membrane-bound organelle in eukaryotes involved in detoxification.
personalized medicine: Use of information on an individual’s genotype to improve its health.
phage: See bacteriophage.
phenocopy: A phenotype induced by an environmental agent (e.g., a temperature shock), which is similar to that produced by a genetic mutation.
phenogram: A branching diagram that links entities by estimates of overall similarity.
phenotype: The observed characteristics of an individual.
phenotypic plasticity: Where the phenotype depends on the environment; see reaction norm.
phenotypic value: The actual value P of a quantitative trait, which is made up of contributions from both genotype and environment (P = G + E).
phenotypic variance (Vp): The variance of the phenotypic value, var(P)= VP.
phosphorite: A sedimentary rock rich in phosphate.
phylogenetic anchor: The use of the phylogeny of a gene to infer the organismal source of a small piece of DNA. It is used in metagenomics.
phylogenetic tree: A diagram showing the evolutionary history of organisms or genes.
phylogenomics: The integration of genome analysis and evolutionary reconstructions into a single composite approach. Also used to refer to the use of phylogenetic trees to predict gene function.
phylogeny: The evolutionary history of organisms or genes.
phylogeography: Inference of population history from the genealogy that connects genes sampled from different geographic locations. It is often based on genealogies inferred from mitochondrial DNA.
phylogram: A phylogenetic tree in which the branch lengths are proportional to the evolutionary distance between nodes. It is also known as an additive tree.
phylotype: The phylogenetic type of an uncultured organism as inferred from analysis of its ribosomal RNA sequence.
physical map: A map that gives the physical location of a genetic variant on the DNA sequence. It contrasts with a genetic map, which is determined using classical genetics.
π: See nucleotide diversity.
placental mammals: See eutherian mammals.
plasmid: A genetic element that can replicate autonomously but is usually smaller than the chromosome. It is frequently required only for specialized conditions and is commonly found in bacteria and archaea and sometimes in eukaryotes.
plastid: A specialized organelle found in plants, algae, and a variety of single-celled eukaryotes. It comes in a variety of forms including chloroplasts and apicoplasts.
plate tectonics: The mechanism by which the plates that make up the surface of the Earth interact with one another, including the formation and subduction of oceanic crust.
pleiotropy: When one allele affects two or more traits.
Pleistocene: The geological period between 1.8 Mya and ~11,000 years ago. It includes major glaciations.
ploidy: The number of copies of each chromosome in the organism.
pluripotency: The ability to differentiate into multiple cell types.
point mutation: A mutation that involves a change from one base to another.
Poisson distribution: The probability that j independent events occur is (λj/j!)e–λ, where λ is the expected number of events.
policing: Where selfish behavior is prevented by other individuals.
polyandrous: Describes females that mate with many males.
polygenic: Influenced by multiple genes.
polymerase chain reaction (PCR): A method for amplifying as little as a single copy of a specific nucleic acid molecule, which is recognized because it binds to a pair of primer sequences.
polymorphism: The presence of multiple alleles or inherited phenotypes at appreciable frequency.
polyphyletic: Describes a group of organisms or genes that do not share a common ancestor.
polyploid: A cell or chromosome carrying more than two copies of each chromosome (e.g., triploid, tetraploid).
polyspermy: Fertilization of an egg by more than one sperm.
polytene chromosome: A chromosome that consists of large numbers of parallel DNA strands, making their structure clearly visible.
polytomy: A portion of a phylogenetic tree in which more than two branches emerge from a single node. It can be used to represent radiation events as well as ambiguities in knowledge.
population bottleneck: A brief reduction in population size, which causes a burst of random genetic drift. It may be caused by a founder effect.
population genetics: Study of the processes that change the genetic composition of populations.
population structure: Any deviation from the ideal state of a single panmictic population. Individuals may be more likely to mate with those in the same place or in the same microhabitat, for example.
positional cloning: Process by which data from genetic crosses are used to identify a DNA fragment that contains a desired gene sequence.
positional homology: When a multiple sequence alignment is used for phylogenetic analysis, residues that are lined up in different sequences are considered to share a common ancestry (i.e., they are derived from a common ancestral residue).
positive selection: See directional selection.
posterior odds ratio: A ratio of the probability of a hypothesis given a particular set of data relative to the probability of another hypothesis given the same data (Prob(H1|D)/Prob(H2|D)). Also known as the odds ratio. This is the value that is estimated in Bayesian phylogenetic reconstruction methods.
postzygotic isolation: Reproductive isolation that acts after production of a hybrid zygote through hybrid inviability or sterility.
power: See statistical power.
prebiotic synthesis: The naturally occurring synthesis of organic compounds before there was life on Earth.
preformation: The view that an embryo develops through the unfolding of preexisting form. This view contrasts with epigenesis.
prenylation: The addition of isoprenoid groups to proteins. It is common in many eukaryotes.
prezygotic isolation: Reproductive isolation that stops production of an F1 zygote by preventing cross-mating.
priapulid: Member of a phylum of worm-like animals (phylum Priapulida).
primary contact: Where populations have been in contact throughout their divergence. It contrasts with secondary contact.
primase: Enzyme used to initiate replication of DNA.
prion: A protein that can take on alternative stable conformations. It is the infectious agent in diseases such as scrapie and bovine spongiform encephalopathy (BSE).
prior odds: A ratio of the probability of one hypothesis relative to the probability of another hypothesis ((Prob(H1)/Prob(H2)), prior to observing any data.
Prisoner’s Dilemma: A game in which both players have a lower fitness when they play the ESS than when they both play the alternative.
probability density: The probability that a random variable is in a small interval of size δx is equal to the probability density multiplied by δx.
probability distribution: A distribution that specifies the chance of every possible outcome; it may be discrete or continuous.
probability of fixation: The chance that a single copy of an allele will ultimately fix throughout the entire population.
prokaryotes: Organisms that do not have nuclei. This includes both bacteria and archaea, two distinct phylogenetic domains of life, and thus the term may represent a polyphyletic grouping of organisms.
proofreading: Correction of DNA replication mistakes by the DNA polymerase enzyme.
protected polymorphism: When each allele will increase from very low frequency, we say that the polymorphism is protected.
protein electrophoresis: A method of analyzing a mixture of proteins by separating the molecules based on physical characteristics such as size, shape, or isoelectric point.
protostome: Member of one of two large groups of bilaterian animals including ecdysozoans and lophotrochozoans (see deuterostome). In this group, the initial embryonic opening becomes the mouth.
pseudogene: A gene that has lost its function and is degenerating under mutation and drift.
psychrophiles: Organisms that prefer to grow at low temperatures.
punctuated equilibrium: A theory devised by Eldredge and Gould (in 1972) based on the observation that in the fossil record species often show long periods of stasis, punctuated by rapid morphological change associated with speciation.
punishment: Where individuals who behave in ways that reduce others’ fitness are made to suffer reduced fitness as a result.
purifying selection: Selection against deleterious alleles.
purines: A class of nucleic acid bases including adenine (A) and guanine (G).
pyrimidines: A class of nucleic acid bases including thymine (T), cytosine (C), and uracil (U).
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
QTL: See quantitative trait locus.
quadratic selection gradient: See selection gradient, quadratic.
quantitative genetics: The study of the inheritance of genetically complex traits.
quantitative trait: Some characteristic that may be influenced by multiple genes and that is studied by the methods of quantitative genetics.
quantitative trait locus (QTL): A region of genome that influences a quantitative trait.
query sequence: A macromolecular sequence (RNA, DNA, or protein) used for searching against a database.
quorum sensing: A mechanism that allows individual bacteria to sense the density of their population.
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
radioisotope dating: See radiometric dating.
radiolarian: Member of a phylum of radially symmetric eukaryotes in the Rhizaria kingdom. They have a strong fossil record.
radiometric dating: A method of dating samples based on analysis of radioactive isotopes and the products of their decay.
random (genetic) drift: The random change in genotype frequency caused by random variation in individual reproduction. Looking backward in time, this process causes coalescence of lineages.
random segregation: During meiosis, the two chromosomes of a pair are distributed randomly to the gametes, each gamete having an equal chance of receiving either chromosome.
random walks: A sequence of random changes; the total change is the sum of all the random steps.
rDNA: See ribosomal DNA.
reaction norm: The set of phenotypes expressed by a single genotype across a range of environments.
recessive: An allele is recessive with respect to a certain phenotype if it produces that phenotype only when present in two copies, that is, as a homozygote.
recessive lethal: See lethal.
reciprocal cross: If a cross is made between A males and B females, then the reciprocal cross is between B males and A females.
reciprocal translocation: A translocation mutation in which parts of two different chromosomes are exchanged.
reciprocally monophyletic: Two groups for which, at every locus, all genes within the group are more closely related to each other than they are to any organisms outside the group. See Web Notes.
recombination: The generation of new combinations of genes.
recombination hot spot: A localized region with exceptionally high recombination rate.
recombination load: The loss of mean fitness caused by recombination breaking up combinations of alleles that are favored by epistasis:
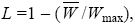
where Wmax is the fitness of the fittest combination of alleles.
recombination, molecular: See molecular recombination.
recombination rate: The proportion of recombinant gametes.
Red Queen: Continual coevolution between two species (e.g., between host and parasite).
reduction principle: If selection is the only process acting, then the recombination rate will tend to decrease.
regression: The way in which a variable y depends on another variable x can be represented by a simple regression model,
y = α + βx + ε,
where ε is a random deviation. β is known as the regression coefficient.
reinforcement: The strengthening of prezygotic isolation through selection against cross-matings that produce unfit hybrid offspring.
relative fitness: See fitness, relative.
relative rate test: A mathematical test for unequal rates of evolution.
replicase: An enzyme that copies any form of genome (i.e., in the origin of life, the genome may not have been DNA or RNA based).
replicative DNA transposon: A DNA-based transposable element that moves itself to a new place in the genome and also leaves a copy in the original location.
replicator: Any entity that can replicate. It usually refers to a DNA-based genome but can include prions and words passed on by cultural transmission.
reproductive assurance: The assurance that an individual can fertilize its eggs or ovules by selfing.
reproductive isolation: The separation of distinct gene pools, as a result of genetic differences that prevent successful interbreeding.
rescue, hybrid: See hybrid rescue allele.
restriction enzyme: An enzyme that cuts DNA at specific sites, typically four to six bases long. The natural function of restriction enzymes is to destroy foreign DNA that enters the cell.
restriction fragment length polymorphism (RFLP): A method for detecting DNA sequence variation that detects variation in the length of fragments cut by restriction enzymes.
retroposon: A DNA transposable element that replicates through an RNA intermediate and does not have long terminal repeats.
retrotransposon: A DNA transposable element that replicates through an RNA intermediate and has long terminal repeats on both sides.
retrovirus: A virus that has an RNA genome and replicates it through a DNA intermediate. DNA can be inserted into the genome of host species.
reverse genetics: Term used to describe any of a variety of molecular methods that allow a wild-type allele of a gene to be targeted and replaced by an engineered mutant allele. See positional cloning.
reverse transcription: Some viruses produce enzymes that reverse the transcription process by copying RNA back into a complementary DNA sequence. This process produces a complementary DNA copy of an RNA molecule and is used by retroviruses and retrotransposons.
RFLP: See restriction fragment length polymorphism.
rhizaria: Another name for the kingdom of eukaryotes known as Cercozoa.
rhyniophyte: Member of an early group of vascular plants.
ribosomal DNA (rDNA): The DNA sequence that codes for the ribosomal RNAs, which form the core of the ribosome.
ribosomal RNA (rRNA): The highly conserved RNA molecules that are found within ribosomes. They are widely used for estimating phylogenies.
ribosome: The protein–RNA complex responsible for translating the genetic code.
ribozyme: An RNA with catalytic activity.
ring species: A chain of interbreeding populations whose ends overlap without interbreeding.
RNA-mediated interference (RNAi): Mechanism of RNA-based regulation of gene function that results from the inhibition of gene expression through the formation of double-stranded RNA.
RNA world: The stage before the evolution of the genetic code when RNA was responsible for both heredity and catalysis.
robust: Used to describe early human fossils—specifically, the large rear teeth of certain Australopithecines. It contrasts with gracile.
robustness: In regards to methods of analysis, a measure of how dependent a result is on the assumptions of the method being used to obtain the result.
rock–paper–scissors game: A game in which strategy A beats B, B beats C, and C beats A.
root: The most ancient branch in a phylogenetic tree.
rRNA: See ribosomal RNA.
runaway process: Arises from the positive feedback between the evolution of a male trait and the female preference for that trait.
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
S: See selection differential.
s: See selection coefficient.
saltation: A variation of large effect; also, a major mutation.
satellite DNA: Highly repeated DNA sequence, which was originally detected as a “satellite” component with a density distinct from the rest of the genome. See microsatellite and minisatellite.
SBT: See shifting balance theory.
secondary contact: Contact between populations that had previously been geographically separate (i.e., allopatric). It contrasts with primary contact.
segregating sites: Sites that are polymorphic in a sample of sequences.
segregation: The movement of two homologous chromosomes during meiosis, one to each pole of the cell. Also, the production of different genotypes in the offspring of heterozygotes, as a result of this random meiotic process.
segregation distortion: Deviation from the expected 1:1 segregation of alleles from a heterozygote at meiosis. It includes both meiotic drive and differential survival of the haploid products of meiosis.
segregation load: The loss of mean fitness caused by the segregation of homozygotes, when polymorphism is maintained by heterozygote advantage:
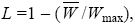
where Wmax is the fitness of the fittest heterozygous genotype.
selection, background: See background selection.
selection coefficient (s): Difference in relative fitness.
selection differential (S): The difference in mean trait value between those that reproduce and the original population.
selection, direct: A change in genotype frequency that is caused by the effects on fitness of the alleles themselves.
selection gradient (β): The gradient of a regression of fitness on trait value.
selection gradient, quadratic (γ): The coefficient of a regression of squared deviation of a trait against fitness. Negative values indicate stabilizing selection, which reduces the variance of the trait.
selectionist: One who holds that most molecular divergence and variation is shaped by selection. It contrasts with neutralist.
selection, purifying: See purifying selection.
selection response: The change in mean trait value over one generation.
selection, truncation: See truncation selection.
selective death: A failure to survive or reproduce; also, a loss of fitness attributable to differences in genotype.
selective sweep: Increase of neutral alleles by hitchhiking with a favorable mutation. This sweeps variation out of the region of the genome surrounding the favorable mutation.
self-fertilization: When a hermaphroditic organism mates with itself.
self-incompatibility: When individuals cannot self-fertilize.
selfing: See self-fertilization.
selfish: A gene that increases its own transmission but reduces the fitness of the individual that carries it.
selfish DNA: Sequences that replicate faster than the rest of the genome and that reduce the fitness of the individual carrying them.
self-replication: Used to describe a molecule or other structure that can cause its own replication.
semiconservative: Describes the replication of double-stranded DNA, where the two new molecules each carry one strand from their parent and a complementary strand that has been newly synthesized.
senescence: See aging.
sensory bias: An innate preference for particular male traits, which did not evolve as a result of the sexual selection caused by that preference.
sequence alignment: A way of arranging the primary sequence of two or more macromolecules (DNA, RNA, or protein) such that individual residues in each sequence are lined up with residues in the other sequence(s). Gaps can be introduced into one or more regions of each sequence to aid in the lining up of residues. Which individual residues are lined up with which other ones depends on the goals of the alignment as well as the methods used to generate the alignment.
sex: Production of offspring that are a mixture between two different parental genotypes.
sex-biased inheritance: Transmission of genes through one or the other sex (e.g., maternal inheritance or sex linkage).
sex chromosome: A chromosome that is inherited differently by the two sexes. In mammals, males carry an X and a Y chromosome, and females carry two X chromosomes. In birds and butterflies, males carry two Z chromosomes, and females carry a Z and a W chromosome.
sex ratio: The ratio of males to females in the population.
sexual selection: Selection arising from variation in the ability to find a mate.
shifting balance theory (SBT): A theory developed by Sewall Wright, in which species evolve toward the best among many alternative adaptive peaks.
short interspersed nucleotide element (SINE): A class of transposable element.
shotgun sequencing: A method of sequencing genomes and environmental samples in which random fragments of DNA are sequenced and then computational methods are used to “reassemble” genomes from the sample.
simple sequence repeats (SSRs): Tandem repeats of a short sequence.
simulated annealing: An optimization algorithm that chooses random changes that improve the desired trait.
SINE: See short interspersed nucleotide element.
single-nucleotide polymorphism (SNP): A SNP (pronounced “snip”) occurs when members of a population vary in which base they carry at a single nucleotide site.
sister chromatid: The two copies of a chromosome after it has been replicated.
site: See nucleotide site.
slime mold: Eukaryotes, from multiple phyla, that normally exist as single-celled amoeba-like organisms but that sometimes gather together into “slugs,” which move together as a unit.
slip-strand mispairing (SSM): A process in which a DNA polymerase adds too many or too few copies of a repetitive sequence during replication.
SNP: See single-nucleotide polymorphism.
Social Darwinism: The idea that, by analogy with natural selection, societies evolve through competition between individuals or groups.
social evolution: The study of the evolutionary consequences of interactions between individuals.
soft selection: Selection where numbers produced by a patch are fixed, independent of the fitness relative survival of the individuals in that patch; see Web Notes.
soma: Those parts of a multicellular organism that will not directly produce gametes. It contrasts with germ line.
spandrels: Triangular spaces formed where two arches intersect. Used by Gould and Lewontin to refer to structures that are necessary by-products of traits that arose for reasons quite unrelated to any present function.
speciation: The process by which new species are formed.
species selection: Selection between species arising from differences in the rate of speciation and/or extinction of lineages.
species tree: A phylogenetic tree showing the relationships among species. It is usually shown to contrast with gene trees, which may include events such as gene duplication and lateral gene transfer.
specificity: Where individual molecules take up a stable conformation with specific biological functions.
sphenopsid: A member of a group of plants that includes trees in the Carboniferous coal swamp forests as well as the living horsetail (Equisetum).
sponges: Common name for members of the phylum Porifera, which are thought to be the earliest branching lineage of animals. Sponges feed by moving water through their bodies using specialized cells with cilia called choanocytes.
sporangium (pl. sporangia): A structure containing spores.
sporophyte: The diploid phase of the life cycle of plants that gives rise to the production of spores by means of meiosis. It contrasts with gametophyte.
SSM: See slip-strand mispairing.
SSR: See simple sequence repeats.
stabilizing selection: Selection that favors intermediate trait values.
standard deviation: Square root of the variance; a measure of the typical magnitude of a random fluctuation.
standard (genetic) code: See canonical code.
standard neutral model: The simplest version of the neutral theory, in which mutation and random genetic drift act in a single panmictic population of constant size.
star genealogy: A genealogy in which all lineages coalesce in a common ancestor at the same time. It is produced by a population bottleneck or by a selective sweep.
statistical power: The chance that the null hypothesis will be rejected when the data are generated by a different model.
stem group: The series of extinct organisms within a clade of living and fossil organisms that lie below the crown group. See crown group.
stem-loop structure: A hairpin structure in an RNA molecule that is maintained by complementary base pairing.
steranes: Chemical derivatives of sterols that have been used as chemical fossils.
stereoisomers: Molecules whose atoms are connected with each other in the same way but are arranged differently in space. This includes enantiomers, which are mirror-image reflections of each other.
sterol: Amphipathic molecules (i.e., they have both hydrophilic and hydrophobic regions) found in the membranes of many organisms, especially eukaryotes.
stigma: The female reproductive organ in a flowering plant, which receives pollen.
strategy: A general term used in game theory. It is equivalent to a phenotype or reaction norm. It refers to the morphology or behavior expressed by an individual under a range of circumstances.
structure: See population structure.
structured coalescent: An extension to the coalescent process in which lineages move from place to place as they trace backward in time while they coalesce.
structured population: See population structure.
style: The long structure between the stigma and ovule in a flower. The pollen must grow through the style in order to fertilize the plant.
substitution: The replacement in a population of one nucleotide or amino acid by another.
substitution load: The total loss of mean fitness caused because favorable alleles substitute gradually by selection rather than instantaneously. It is the integral of
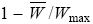
over time, where the mean fitness 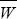 increases toward the maximum fitness Wmax. increases toward the maximum fitness Wmax.
substitution matrix: A table showing the probability of change occurring between different macromolecular residues (nucleotides or amino acids).
subtree pruning and regrafting: A method for searching phylogenetic tree space whereby a new tree is generated from a starting tree by moving entire branches (along with subbranches) to a new position in the tree.
supercoiling: Higher-order twisting of DNA strands.
supergene: A cluster of tightly linked genes, which allow distinct alternative morphs to be maintained as a polymorphism within one population.
suppressor mutation: A secondary mutation that can cancel the effect of a primary mutation, resulting in a wild-type phenotype.
symbiosis: A close association between two organisms.
sympatric speciation: The separation of a single population into two or more reproductively isolated species in the absence of any geographic barriers.
sympatry: Coexistence in the same place.
synapsis: The lining up of homologous chromosomes in meiosis.
syngameon: A botanical term, referring to a cluster of taxa that are morphologically distinct and yet exchange genes.
syngamy: The union of two genomes, which leads to a doubling of ploidy level.
synonymous mutation: A point mutation in a protein-coding region that changes a codon such that it does not alter the resulting amino acid sequence of the protein.
syntax: The rules that determine how words combine to make phrases and sentences.
systematics: See taxonomy.
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
tagging single-nucleotide polymorphisms (tSNPs): A set of single-nucleotide polymorphisms that distinguish different haplotypes segregating in a population.
tandem (DNA) repeats: A series of repeated sequences, arrayed next to each other.
tandem duplication: A duplication mutation in which the duplicated DNA is found next to the original DNA.
tautomerization: The spontaneous isomerization of a nitrogen base from its normal keto (or amino) form to an alternative hydrogen-bonding enol (or imino) form.
taxon (pl. taxa): A unit of classification (e.g., species, genus).
taxonomy: Classification of living organisms (also known as systematics).
TDT: See transmission disequilibrium test.
telocentric: A chromosome with the centromere in the middle.
telomerase: An enzyme complex found in most eukaryotes that maintains the length of telomeres through successive divisions.
telomere: The end of a chromosome.
tetraploid: Carrying four copies of each chromosome gene.
theistic evolution: A view in which religious teachings are seen as compatible with biological evolution. Species share common ancestry and change through natural selection and other evolutionary processes but with divine guidance and intervention.
theory: A set of interconnected hypotheses that leads to testable predictions.
thermophile: An organism with a growth temperature optimum between 50°C and 80°C.
θ: The product 4Neμ, which measures the relative rates of random drift (1/2Ne) and mutation (μ) and hence determines the variation maintained in a balance between these processes.
threshold model: A model that states that a discrete trait is present only when the quantity of an underlying continuous trait is greater than some threshold.
tip: The terminal node on a phylogenetic tree.
tit-for-tat: The winning strategy in the repeated Prisoner’s Dilemma game.
tracheid: A cell with strengthened walls that functions to transport fluid within plants.
trade-off: A situation where one trait cannot be increased without decreasing another. The term is used to describe constraints in optimization arguments.
tragedy of the commons: Where self-interested exploitation of common resources leads to a worse outcome for all. See Prisoner’s Dilemma.
trait: See quantitative trait.
transcription: Replication of an RNA strand complementary to a DNA sequence.
transcription factor: A molecule that binds to the promoter and regulates transcription.
transduction: The movement of genes from a donor cell to a recipient cell with a virus as the vector.
transfer RNA (tRNA): An RNA molecule that couples a specific amino acid to a specific sequence of three bases. It is responsible for translating the genetic code.
transformation: The introduction of a fragment of DNA into a genome. Transformation occurs naturally in some bacteria and archaea; in the laboratory, it is the basis of genetic engineering.
transgenic: Genetically manipulated to carry genes from another individual or species.
transition: A mutation in which a purine replaces another purine or a pyrimidine replaces another pyrimidine.
translation: Synthesis of protein with amino acid sequence encoded by an RNA sequence.
translocation: A rearrangement mutation in which part of one chromosome breaks away and joins another.
transmission disequilibrium test (TDT): A statistical test that detects associations between genetic markers and disease alleles by looking for marker alleles that are transmitted in excess to affected offspring.
transposable element: A genetic element that can move from one location in the genome to another.
transposon: See transposable element.
transversion: A mutation in which a pyrimidine replaces a purine, or vice versa.
Tree of Life: A phylogenetic tree showing the relationships among all cellular organisms.
trigonotarbid: Member of an order of extinct terrestrial spider-like animals (order Trigonotarbida).
trimerophyte: Member of an early group of vascular plants.
triploid: Carrying three genomes.
trisomy: Possession of three copies of one chromosome (in humans this is abnormal because there are normally only two copies).
tristyly: A polymorphism with three different arrangements of anther and stigma. It promotes outcrossing.
tRNA: See transfer RNA.
trochophore: Larval type characteristic of many protostomes including annelids and many mollusks.
true breeding: A population or individual that produces genetically identical offspring.
truncation selection: Selection that eliminates those with the largest (or smallest) trait values.
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
UEP: See unique event polymorphism.
ultracentrifuge: A very high-speed centrifuge used to separate macromolecules.
ultrameric tree: A phylogenetic tree in which the branch lengths are constrained to all be equidistant from the root. It is also known as a dendrogram.
uncultured microbes: Microbes that have never successfully been grown in isolation in the laboratory.
underdominance: Describes heterozygotes that have lower trait values (usually lower fitness) than either homozygote.
unequal crossover: The outcome when two tandemly repeated sequences do not pair correctly. A crossover between them produces one genome with fewer repeats and one with more.
uniformitarianism: The assumption that the same natural processes acted in the past as are observed to operate now.
unique event polymorphism (UEP): An allele for which all copies derive from a single mutational event.
universal genetic code: See canonical code.
universal homology: A homologous trait found in all cellular organisms.
unrooted tree: A phylogenetic tree in which the root is not shown (frequently because it is not known).
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
VA: See additive genetic variance.
variable number tandem repeat (VNTR): A general term for tandemly repeated sequences. It includes satellite, microsatellite, and minisatellite sequences.
variables: Quantities that describe the state of a system and that evolve through time. Contrast with parameters.
variance: The mean squared deviation from the average:
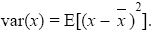
variance component: A contribution to the phenotypic variance due to a specific kind of variation. It includes environmental, additive, dominance, and interaction variances.
var(x): See variance.
vascular plants: Division of plants with vascular tissues, which function in transporting fluids.
VD: See dominance variance.
VE: See environmental variance.
vertical descent: The evolution of species by a branching pattern.
vertical evolution: See vertical descent.
vertical inheritance: The transmission of traits from parent to offspring.
vertical transmission: See vertical inheritance.
VG: See genotypic variance.
virulence: The degree of pathogenicity of a parasite.
VM: See mutational variance.
VNTR: See variable number tandem repeat.
VW: See additive genetic variance in fitness.
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
wave of advance: A favorable allele advances behind a moving cline, known as a wave of advance.
wild type: The commonest allele at a locus or the most common genotype.
wobble pairing: The ability of tRNAs to hybridize with codons even when only the first two nucleotides follow the standard G-C and A-U base-pairing rules.
Wright–Castle estimator: A method for estimating the number of genes that influence a quantitative trait. It is based on a comparison between the variance in an F2 population and the difference in means between the parent populations.
Wright–Fisher model: A standard model of random genetic drift, in which each gene is drawn at random from 2N genes in the previous generation.
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
xerophiles: Organisms that prefer to grow in very dry conditions.
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
P
Q
R
S
T
U
V
W
X
Y
Z
zosterophyll: A type of early vascular plant that carried spores laterally along the stem.
zygote: The diploid cell formed by union of two haploid gametes.
|

