|
Problem 22.1
A species sometimes interbreeds with another species; on average, 10% of matings are with the other species. The F1 offspring from such matings have normal viability but are completely sterile. Offspring spend their early life together, with each family growing within its own patch. There is intense competition within patches, so that the same number emerges from each.
|
| **i) |
Assuming that all the fertile offspring in a patch have the same father, how rapidly will a rare dominant allele that causes the early death of F1 hybrids increase?
|
| *ii) |
How rapidly will it increase if all the offspring in a family have different fathers (i.e., soft selection)? HINT 22A
|
|
Problem 22.2
Two species diverged 1 Mya, but they have since hybridized at a constant rate: In each species, a fraction m = 5 × 10–7 of breeding individuals come from the other species. The effective population sizes of each species and of the ancestral species are all Ne = 105. Individuals reproduce annually, and so a year equals one generation. See Figure P22.1.
|
| *i) |
What is the chance that two ancestral lineages, sampled from different species, both trace back into the ancestral population without ever crossing between the species? Given that this happens, what is their expected time to coalescence?
|
| **ii) |
What is the chance that one of the lineages crosses into the other species but that, nevertheless, the lineages both trace back into the ancestral population? HINT 22B
|
| **iii) |
What is the chance that two lineages coalesced recently—within the past 105 generations, say?
|
| ***iv) |
If many loci were surveyed, what would the distribution of coalescence times look like? HINT 22C
|
|
Problem 22.3
Approximately 9.3 Mb of autosomal sequences from human, chimpanzee, and gorilla were aligned; sequences from orangutan and macaque were also aligned to provide an outgroup. Sites were classified according to whether variants were found only in human (H), only in chimpanzee (C), only in chimpanzee and human (HC), and so on. The total numbers of sites were as follows:

These numbers have been corrected for multiple hits (see Chapter 27); so you can assume that these represent mutations that occurred only once in the genealogy.
The simplest hypothesis to account for these data is that the population ancestral to humans and chimpanzees separated T0 years ago and that the population ancestral to gorillas split off from the ancestors of humans and chimpanzees a time T1 before that. Assume that both ancestral populations had effective size Ne. Figure P22.2A shows an example of where the genealogy at a site (thin lines) is the same as the species’ phylogeny; the star marks a mutation ancestral to H and C, which would appear in class HC. Figure P22.2B shows an example where C and G are most closely related, which disagrees with the species’ phylogeny; the asterisk marks a mutation that would appear in class CG. This phenomenon is known as lineage sorting (see p. 628).
|
| **i) |
Explain what features of the data favor the species’ phylogeny shown in Figure P22.2A,B.
|
| **ii) |
What is the total number of sites that differ between human and chimpanzee? Between human and gorilla? Between chimpanzee and gorilla? Use these numbers to estimate the time T1 between the two speciation events. (Assume a mutation rate of μ = 10–8 per site per generation.)
|
| ***iii) |
What is the chance that the lineages ancestral to humans and to chimpanzees do not coalesce during the time, T1, between the speciation events? Given that they did not coalesce, what is the chance that the next coalescence event is with the gorilla lineage, so that the genealogy is different from the phylogeny (e.g., as in Fig. P22.2B, rather than Fig. 22.2A)?
|
| ***iv) |
What is the expected number of sites in the class CG or HG? HINT 22D
|
| ***v) |
Use the answers to iii) and iv) to estimate the effective size of the ancestral population, the fraction of the genome that has a genealogy different from the phylogeny, and the time when humans and chimpanzees formed separate species. NOTE 22B
|
|
Problem 22.4
A simple model for the evolution of reproductive isolation in allopatry is that each population accumulates substitutions at a steady rate. These substitutions might occur by random drift or because they are positively selected, but they do not lead to any incompatibilities being expressed during the course of evolution within the diverging populations. Substitutions can occur at very many sites, and so we can assume that the substitutions in each population are different. There is a small chance p that any pair of alleles will cause incompatibility; each incompatibility that is expressed in a hybrid reduces its fitness by a factor (1 – s), where s = 10%. For example, if each population accumulates 100 substitutions, then there are 100 × 100 = 10,000 pairs of alleles that might prove incompatible. If p = 10–3, then we expect 100 × 100 × p = 10 incompatibilities to arise, and we expect the fitness of hybrids to be reduced by a factor ~(1 – s)10 = 0.35.
Alleles might be dominant or recessive. Thus, if there is an incompatibility between two dominant alleles, A1 and B1, then the F1 genotype A1B0/A0B1 will suffer, as will the three other genotypes A1B0/A1B1, A1B1/A0B1, A1B1/A1B1, which could appear in F2. On the other hand, if A1 and B1 are both recessive, then only the double homozygote A1B1/A1B1 will express the incompatibility. Assume that most alleles are recessive, with only a proportion α = 0.01 of alleles being dominant; assume that p = 10–3, as before. NOTE 22C
|
| *i) |
Suppose that 2000 substitutions have occurred at autosomal genes in each of two diverging lineages. By how much will the fitness of an F1 hybrid be reduced by incompatibilities between these alleles? HINT 22E
|
| *ii) |
Now, suppose that 500 substitutions have occurred on the X chromosome. By how much will the fitnesses of F1 females be reduced?
|
| **iii) |
By how much will the fitness of F1 males be reduced? HINT 22F
|
| **iv) |
Sketch a graph of the fitnesses of F1 males and F1 females against the total number of substitutions in each lineage. (Assume that on average a fraction χ = 0.2 of substitutions are on the X, rather than on the autosomes.)
|
| ***v) |
What will be the average fitness of F2 males and females, for the case of 200 autosomal and 50 X-linked substitutions in each lineage, and with α = 0.01 of the incompatibilities dominant, as before?
|
|
Problem 22.5
The chance that a single copy of a chromosome rearrangement will fix in a population with effective size Ne, despite selection s against heterozygotes, is close to
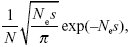
where N is the actual number of individuals. NOTE 22D
|
| *i) |
If the rate of mutation to chromosome rearrangements is μ = 10–4 per haploid genome per generation, and the selection against heterozygotes is s ~ 10%, what is the rate at which the chromosome arrangement changes in demes of effective size Ne = 10, 20, and 30? Sketch a graph of this rate of chromosomal evolution against deme size. HINT 22G
|
| **ii) |
Once a chromosome rearrangement is fixed within a small deme, how does it spread through the whole species? If demes become extinct every so often and are recolonized by individuals from another deme, then the new chromosome arrangement may spread. At what rate does the whole species change? HINT 22H
|
| **iii) |
What factors might increase the rate of chromosomal evolution? NOTE 22E
|
|
Problem 22.6
Under the simple model of Problem 22.5, the rate of chromosomal evolution is faster for more weakly selected changes, but, on the other hand, such changes contribute little to isolation.
|
| **i) |
Sketch a graph of the rate of evolution of reproductive isolation plotted against the selection coefficient s for Ne = 40, and μ = 10–4.
|
| *ii) |
For what selection coefficient does isolation evolve most rapidly? HINT 22I
|
|
Problem 22.7
The Dobzhansky–Muller model allows incompatibilities to evolve in allopatry, without being opposed by selection and without any difference in environment. Can such incompatibilities be maintained after divergent populations come back into contact?
To keep the model as simple as possible, we consider two unlinked genes (A, B) in a haploid population. In one population, an allele AP increases relative fitness by s = 0.01 and replaces allele AQ. In another population, an allele BP replaces BQ, with the same selective advantage. However, APBP genotypes die. You can assume that there is no association between the genes (i.e., they are in linkage equilibrium). NOTE 22F
A population fixed for APBQ receives a fraction m of its individuals from a population fixed for the opposite genotype, AQBP. As a result, booth genes are polymorphic, with pA < 1 and 0 < pB.
|
| *i) |
What are the average fitnesses of the AP, AQ alleles? What is the selective advantage of AP over AQ? Similarly, what are the average fitnesses of the BP, BQ alleles and the selective advantage of BP over BQ? HINT 22J
|
| **ii) |
Write down equations for the change in allele frequency due to selection and migration. HINT 22K
|
| ***iii) |
What are the equilibria, for migration rates m = 0.002 and m = 0.0035? HINT 22L
|
| ***iv) |
What is the threshold migration rate above which divergence collapses? How does this compare with the threshold in the simple case, with no incompatibility, where gene flow brings in alleles that are locally disadvantageous? (See Box 28.3.)
|
|
Problem 22.8
In Problem 22.7 we asked whether incompatibilities could persist despite gene flow. Here, we ask whether they could arise in the first place despite gene flow. Can incompatibilities evolve under the Dobzhansky–Muller model when two populations are connected by gene flow? As in Problem 22.4, assume that substitutions can occur at very many sites and that there is a small chance p = 10–3 that any pair of substitutions are incompatible. Imagine two populations of N = 104 diploid individuals that exchange genes at a very low rate, m = 10–6 per generation, and that share the same environment. Favorable alleles arise and fix at a rate Λ = 0.02 per generation within each of the two populations. A new allele that fixes in one population will eventually spread to fix in the other, unless there is another allele there that is incompatible with it. Thus, if a stable genetic difference is to be established, then either the new allele has to be incompatible with a divergent allele that is already established in the other population or an incompatible allele has to fix in the other population before the new allele gets transmitted across.
|
| **i) |
Suppose that an allele with advantage s = 0.05 has fixed in one population. Assuming that it does not meet another, incompatible, allele, what is the chance per generation that it will be successfully introduced into the other population and will increase to fixation there? What is the expected time before this occurs?
|
| **ii) |
What is the chance that an incompatible allele will arise in the other population before the new allele can get established there and so prevent it from invading?
|
| *iii) |
If there are already k differences between the populations, there is a chance that the new allele will be incompatible with one of these. What is this chance?
|
| ***iv) |
*C28* Write down a differential equation for the rate of increase of the number of differences. Use this to find how long it will take for 100 incompatibilities to evolve.
|
| **v) |
What factors could aid divergence?
|
Lande R. 1979. Effective deme sizes during long-term evolution estimated from rates of chromosomal rearrangement. Evolution 33: 234–251.
Wright S. 1941. On the probability of fixation of reciprocal translocations. Am. Nat. 75: 513–522.

